Analysis and figures¶
for Tests 1-5
[16]:
%load_ext autoreload
%autoreload 2
import logging
import os
import pathlib
import pickle
import string
import matplotlib as mpl
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import PIL
import pygmt
import seaborn as sns
import verde as vd
import xarray as xr
import xrft
from invert4geom import plotting, utils
from invert4geom import synthetic as inv_synthetic
from polartoolkit import fetch, maps, profiles
from polartoolkit import utils as polar_utils
from synthetic_bathymetry_inversion import ice_shelf_stats, synthetic
sns.set_theme()
os.environ["POLARTOOLKIT_HEMISPHERE"] = "south"
logging.getLogger().setLevel(logging.INFO)
The autoreload extension is already loaded. To reload it, use:
%reload_ext autoreload
[17]:
# set grid parameters
spacing = 2e3
inversion_region = (-40e3, 110e3, -1600e3, -1400e3)
plot_region = inversion_region
[2]:
true_density_contrast = 1476
bathymetry, basement, _ = synthetic.load_synthetic_model(
spacing=spacing,
inversion_region=inversion_region,
buffer=spacing * 10,
basement=True,
zref=0,
bathymetry_density_contrast=true_density_contrast,
plot_topography=False,
plot_gravity=False,
)
buffer_region = polar_utils.get_grid_info(bathymetry)[1]
# clip to plot region
bathymetry = bathymetry.sel(
easting=slice(plot_region[0], plot_region[1]),
northing=slice(plot_region[2], plot_region[3]),
)
basement = basement.sel(
easting=slice(plot_region[0], plot_region[1]),
northing=slice(plot_region[2], plot_region[3]),
)
requested spacing (2000.0) is smaller than the original (5000.0).
requested spacing (2000.0) is smaller than the original (5000.0).
Load results of notebooks 01-05¶
[3]:
damping_studies = []
density_studies = []
optimal_results = []
# uncertainty_results = []
constraint_points_dfs = []
sensitivity_results = []
final_bathymetries = []
final_uncertainties = []
final_stochastic_means = []
inversion_titles = [
"Best case scenario",
"Errors in gravity",
"Airborne survey",
"Regional gravity",
"Realistic scenario",
]
for i in range(1, 6):
# get names for various studies / results
damping_study_fname = (
f"../results/Ross_Sea/Ross_Sea_0{i}_damping_cv_damping_cv_study.pickle"
)
if i >= 4:
density_study_fname = (
f"../results/Ross_Sea/Ross_Sea_0{i}_density_cv_kfolds_study.pickle"
)
else:
density_study_fname = (
f"../results/Ross_Sea/Ross_Sea_0{i}_density_cv_zref_density_cv_study.pickle"
)
optimal_results_fname = f"../results/Ross_Sea/Ross_Sea_0{i}_optimal_results.pickle"
# open studies
with pathlib.Path(damping_study_fname).open("rb") as f:
damp_study = pickle.load(f)
with pathlib.Path(density_study_fname).open("rb") as f:
dens_study = pickle.load(f)
with pathlib.Path(optimal_results_fname).open("rb") as f:
optimal_result = pickle.load(f)
topo_results, _, _, _ = optimal_result
final_bathymetry = topo_results.set_index(["northing", "easting"]).to_xarray().topo
uncertainty_ds = xr.load_dataset(
f"../results/Ross_Sea/Ross_Sea_0{i}_uncertainty.nc"
)
sensitivity_ds = xr.load_dataset(
f"../results/Ross_Sea/Ross_Sea_0{i}_sensitivity.nc"
)
# clip to inversion region
final_bathymetry = final_bathymetry.sel(
easting=slice(plot_region[0], plot_region[1]),
northing=slice(plot_region[2], plot_region[3]),
)
final_uncert = sensitivity_ds.full_stdev.sel(
easting=slice(plot_region[0], plot_region[1]),
northing=slice(plot_region[2], plot_region[3]),
)
final_stochastic_mean = uncertainty_ds.weighted_mean.sel(
easting=slice(plot_region[0], plot_region[1]),
northing=slice(plot_region[2], plot_region[3]),
)
# add to lists
damping_studies.append(damp_study)
density_studies.append(dens_study)
optimal_results.append(optimal_result)
constraint_points_dfs.append(
pd.read_csv(f"../results/Ross_Sea/Ross_Sea_0{i}_constraint_points.csv")
)
sensitivity_results.append(
xr.load_dataset(f"../results/Ross_Sea/Ross_Sea_0{i}_sensitivity.nc")
)
final_bathymetries.append(final_bathymetry)
final_stochastic_means.append(final_stochastic_mean)
final_uncertainties.append(final_uncert)
[4]:
abs_inversion_errors = []
uncert_errors = []
for i, g in enumerate(final_bathymetries):
abs_inversion_error = np.abs(bathymetry - g)
abs_inversion_errors.append(abs_inversion_error)
uncert_errors.append(abs_inversion_error - final_uncertainties[i])
[5]:
for i, t in enumerate(inversion_titles):
print(i + 1, t)
starting_bathymetry = (
optimal_results[i][0]
.set_index(["northing", "easting"])
.to_xarray()
.starting_topo
)
starting_bathymetry = starting_bathymetry.sel(
easting=slice(plot_region[0], plot_region[1]),
northing=slice(plot_region[2], plot_region[3]),
)
print(
f"\tStarting bathymetry RMSE: {round(utils.rmse(bathymetry - starting_bathymetry), 2)} m"
)
print(
f"\tDeterministic bathymetry RMSE: {round(utils.rmse(bathymetry - final_bathymetries[i]), 2)} m"
)
print(
f"\tDeterministic bathymetry max absolute error: {round(vd.maxabs(bathymetry - final_bathymetries[i]), 2)} m"
)
print(
f"\tStochastic bathymetry RMSE: {round(utils.rmse(bathymetry - final_stochastic_means[i]), 2)} m"
)
print(f"\tUncertainty RMS: {round(utils.rmse(final_uncertainties[i]), 2)} m")
print(f"\tUncertainty RMSE: {round(utils.rmse(uncert_errors[i]), 2)} m")
scores = damping_studies[i].trials_dataframe().value.to_numpy()
parameters = damping_studies[i].trials_dataframe().params_damping.to_numpy()
param_name = "Damping"
df0 = pd.DataFrame({"scores": scores, "parameters": parameters})
df = df0.sort_values(by="parameters")
best = df.scores.argmin()
best_damping = df.parameters.to_numpy()[best]
print(f"\tOptimal damping: {round(best_damping, 4)}")
scores = density_studies[i].trials_dataframe().value.to_numpy()
parameters = (
density_studies[i].trials_dataframe().params_density_contrast.to_numpy()
)
param_name = "Density"
df0 = pd.DataFrame({"scores": scores, "parameters": parameters})
df = df0.sort_values(by="parameters")
best = df.scores.argmin()
best_density = df.parameters.to_numpy()[best]
print(f"\tOptimal density: {round(best_density)} kg/m^3")
print(f"\tDensity contrast error: {best_density - true_density_contrast} kg/m^3")
constraint_points = constraint_points_dfs[i]
rmse = utils.rmse(
constraint_points.starting_bathymetry - constraint_points.inverted_topography
)
print(f"\tConstraint points RMSE: {round(rmse, 3)} m")
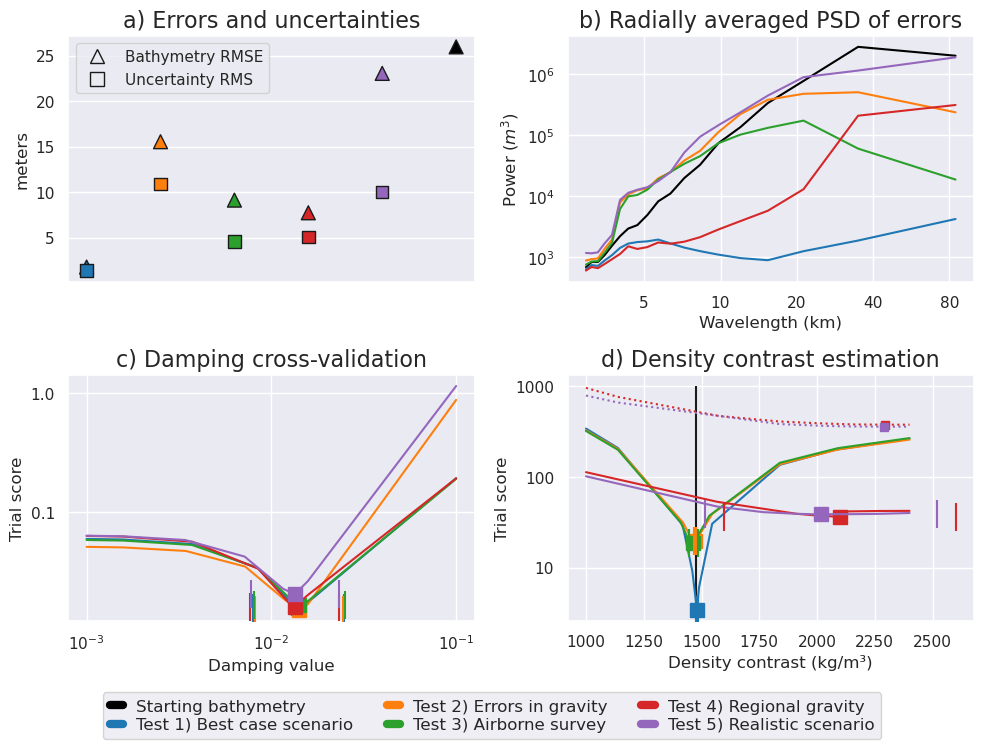
1 Best case scenario
Starting bathymetry RMSE: 25.98 m
Deterministic bathymetry RMSE: 1.76 m
Deterministic bathymetry max absolute error: 24.87 m
Stochastic bathymetry RMSE: 2.05 m
Uncertainty RMS: 1.37 m
Uncertainty RMSE: 1.36 m
Optimal damping: 0.014
Optimal density: 1480 kg/m^3
Density contrast error: 4 kg/m^3
Constraint points RMSE: 3.381 m
2 Errors in gravity
Starting bathymetry RMSE: 25.98 m
Deterministic bathymetry RMSE: 15.54 m
Deterministic bathymetry max absolute error: 132.86 m
Stochastic bathymetry RMSE: 15.91 m
Uncertainty RMS: 10.91 m
Uncertainty RMSE: 9.86 m
Optimal damping: 0.0141
Optimal density: 1471 kg/m^3
Density contrast error: -5 kg/m^3
Constraint points RMSE: 19.532 m
3 Airborne survey
Starting bathymetry RMSE: 25.98 m
Deterministic bathymetry RMSE: 9.12 m
Deterministic bathymetry max absolute error: 148.02 m
Stochastic bathymetry RMSE: 8.9 m
Uncertainty RMS: 4.56 m
Uncertainty RMSE: 6.4 m
Optimal damping: 0.0142
Optimal density: 1464 kg/m^3
Density contrast error: -12 kg/m^3
Constraint points RMSE: 19.212 m
4 Regional gravity
Starting bathymetry RMSE: 25.98 m
Deterministic bathymetry RMSE: 7.73 m
Deterministic bathymetry max absolute error: 31.53 m
Stochastic bathymetry RMSE: 7.75 m
Uncertainty RMS: 5.07 m
Uncertainty RMSE: 5.84 m
Optimal damping: 0.0134
Optimal density: 2099 kg/m^3
Density contrast error: 623 kg/m^3
Constraint points RMSE: 3.538 m
5 Realistic scenario
Starting bathymetry RMSE: 25.98 m
Deterministic bathymetry RMSE: 23.06 m
Deterministic bathymetry max absolute error: 109.49 m
Stochastic bathymetry RMSE: 20.49 m
Uncertainty RMS: 10.04 m
Uncertainty RMSE: 16.17 m
Optimal damping: 0.0134
Optimal density: 2017 kg/m^3
Density contrast error: 541 kg/m^3
Constraint points RMSE: 17.707 m
[7]:
starting_bathymetry_no_errors = (
optimal_results[0][0].set_index(["northing", "easting"]).to_xarray().starting_topo
)
starting_bathymetry_no_errors = starting_bathymetry_no_errors.sel(
easting=slice(plot_region[0], plot_region[1]),
northing=slice(plot_region[2], plot_region[3]),
)
[8]:
print(
f"Starting bathymetry MAE: {np.abs(bathymetry - starting_bathymetry_no_errors).mean().to_numpy()}"
f"\nStarting bathymetry RMSE: {utils.rmse(bathymetry - starting_bathymetry_no_errors)}"
f"\nmax absolute error: {np.abs(bathymetry - starting_bathymetry_no_errors).max().to_numpy()}"
)
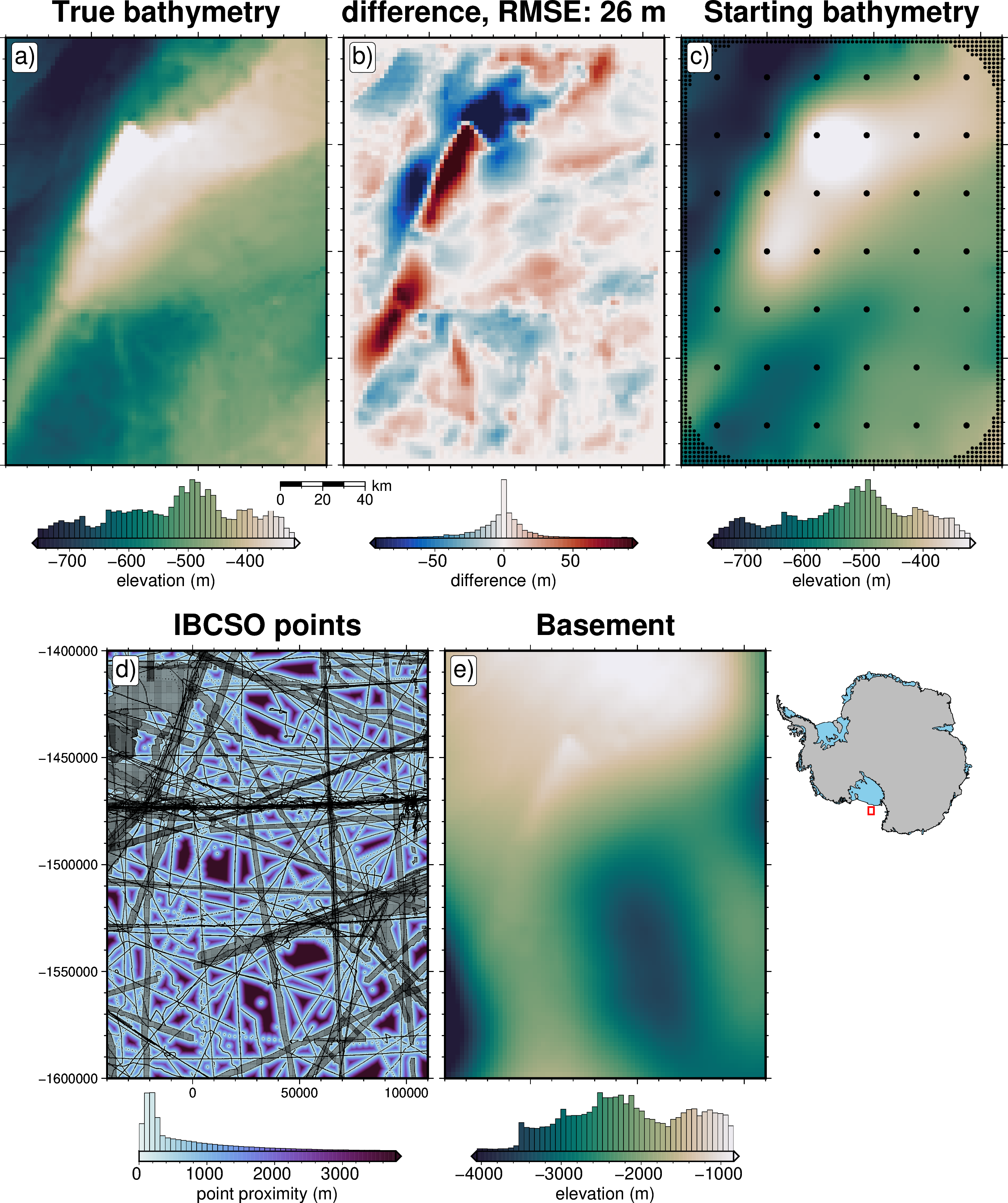
Starting bathymetry MAE: 15.0527279185003
Starting bathymetry RMSE: 25.97734942125003
max absolute error: 164.39923609925518
Location figure¶
[8]:
ibcso_points_gdf, ibcso_polygons_gdf = fetch.ibcso_coverage(
region=plot_region,
)
[9]:
ibcso_points_gdf
[9]:
| dataset_name | dataset_tid | weight | geometry | easting | northing | |
|---|---|---|---|---|---|---|
| 3 | NBP94_6.xyz | 10 | 10 | POINT (109770.251 -1589039.712) | 109770.251441 | -1.589040e+06 |
| 3 | NBP94_6.xyz | 10 | 10 | POINT (109541.648 -1588891.732) | 109541.648463 | -1.588892e+06 |
| 3 | NBP94_6.xyz | 10 | 10 | POINT (109295.696 -1588705.992) | 109295.696152 | -1.588706e+06 |
| 3 | NBP94_6.xyz | 10 | 10 | POINT (109042.6 -1588513.108) | 109042.599997 | -1.588513e+06 |
| 3 | NBP94_6.xyz | 10 | 10 | POINT (108798.689 -1588330.43) | 108798.688784 | -1.588330e+06 |
| ... | ... | ... | ... | ... | ... | ... |
| 53 | NGA_various.xyz | 13 | 5 | POINT (46074.726 -1401160.721) | 46074.726123 | -1.401161e+06 |
| 36 | NBP93_8.xyz | 10 | 10 | POINT (47246.316 -1401134.187) | 47246.316386 | -1.401134e+06 |
| 37 | NBP93_7.xyz | 10 | 10 | POINT (47246.316 -1401134.187) | 47246.316386 | -1.401134e+06 |
| 36 | NBP93_8.xyz | 10 | 10 | POINT (47393.275 -1401065.81) | 47393.275443 | -1.401066e+06 |
| 37 | NBP93_7.xyz | 10 | 10 | POINT (47393.275 -1401065.81) | 47393.275443 | -1.401066e+06 |
108983 rows × 6 columns
[10]:
polygon_points = ice_shelf_stats.polygons_to_points(
ibcso_polygons_gdf,
spacing=500,
)
polygon_points
[10]:
| geometry | easting | northing | |
|---|---|---|---|
| 0 | POINT (-39500 -1509160.059) | -39500.000000 | -1.509160e+06 |
| 1 | POINT (-39500 -1508660.059) | -39500.000000 | -1.508660e+06 |
| 2 | POINT (-39500 -1508160.059) | -39500.000000 | -1.508160e+06 |
| 3 | POINT (-39500 -1507660.059) | -39500.000000 | -1.507660e+06 |
| 4 | POINT (-39500 -1507160.059) | -39500.000000 | -1.507160e+06 |
| ... | ... | ... | ... |
| 9870 | POINT (-38380.519 -1504416.675) | -38380.519348 | -1.504417e+06 |
| 9871 | POINT (-38785.39 -1504419.603) | -38785.389511 | -1.504420e+06 |
| 9872 | POINT (-39190.26 -1504422.531) | -39190.259674 | -1.504423e+06 |
| 9873 | POINT (-39595.13 -1504425.458) | -39595.129837 | -1.504425e+06 |
| 9874 | POINT (-40000 -1504428.386) | -40000.000000 | -1.504428e+06 |
67682 rows × 3 columns
[11]:
total_ibcso_points = pd.concat([ibcso_points_gdf, polygon_points])
total_ibcso_points
[11]:
| dataset_name | dataset_tid | weight | geometry | easting | northing | |
|---|---|---|---|---|---|---|
| 3 | NBP94_6.xyz | 10.0 | 10.0 | POINT (109770.251 -1589039.712) | 109770.251441 | -1.589040e+06 |
| 3 | NBP94_6.xyz | 10.0 | 10.0 | POINT (109541.648 -1588891.732) | 109541.648463 | -1.588892e+06 |
| 3 | NBP94_6.xyz | 10.0 | 10.0 | POINT (109295.696 -1588705.992) | 109295.696152 | -1.588706e+06 |
| 3 | NBP94_6.xyz | 10.0 | 10.0 | POINT (109042.6 -1588513.108) | 109042.599997 | -1.588513e+06 |
| 3 | NBP94_6.xyz | 10.0 | 10.0 | POINT (108798.689 -1588330.43) | 108798.688784 | -1.588330e+06 |
| ... | ... | ... | ... | ... | ... | ... |
| 9870 | NaN | NaN | NaN | POINT (-38380.519 -1504416.675) | -38380.519348 | -1.504417e+06 |
| 9871 | NaN | NaN | NaN | POINT (-38785.39 -1504419.603) | -38785.389511 | -1.504420e+06 |
| 9872 | NaN | NaN | NaN | POINT (-39190.26 -1504422.531) | -39190.259674 | -1.504423e+06 |
| 9873 | NaN | NaN | NaN | POINT (-39595.13 -1504425.458) | -39595.129837 | -1.504425e+06 |
| 9874 | NaN | NaN | NaN | POINT (-40000 -1504428.386) | -40000.000000 | -1.504428e+06 |
176665 rows × 6 columns
[12]:
fig = maps.basemap(region=plot_region, points=total_ibcso_points, points_style="p2p")
fig.plot(
ibcso_polygons_gdf,
fill="cyan",
close=True,
transparency=50,
)
fig.show()
[13]:
# calculate gravity data median distance
grd = fetch.resample_grid(bathymetry, spacing=100, verbose="q").rename(
{"x": "easting", "y": "northing"}
)
min_dist = utils.dist_nearest_points(
total_ibcso_points,
grd,
).min_dist
print(f"mean minimum distance: {int(min_dist.mean().to_numpy())} m")
print(f"median minimum distance: {int(min_dist.median().to_numpy())} m")
requested spacing (100) is smaller than the original (2000.0).
mean minimum distance: 788 m
median minimum distance: 359 m
[14]:
####
# Bathymetry
####
bathy_lims = polar_utils.get_combined_min_max(
[bathymetry, starting_bathymetry_no_errors], robust=True
)
fig = maps.plot_grd(
bathymetry,
fig_height=10,
cmap="rain",
reverse_cpt=True,
cpt_lims=bathy_lims,
title="True bathymetry",
cbar_label="elevation (m)",
cbar_font="18p,Helvetica,black",
frame=["nsew", "xaf10000", "yaf10000"],
hist=True,
scalebar=True,
scalebar_position="jBR+o-.9c/-.6c",
)
fig.text(
position="TL",
justify="TL",
text="a)",
font="16p,Helvetica,black",
offset="j.2c",
no_clip=True,
fill="white",
clearance="+tO",
pen=True,
)
####
# Difference with starting
####
dif = bathymetry - starting_bathymetry_no_errors
fig = maps.plot_grd(
dif,
fig=fig,
origin_shift="x",
region=plot_region,
title=f"difference, RMSE: {round(utils.rmse(dif))} m",
cbar_label="difference (m)",
cbar_font="18p,Helvetica,black",
cmap="balance+h0",
cpt_lims=polar_utils.get_min_max(
dif, robust=True, absolute=True, robust_percentiles=(0.01, 0.99)
),
frame=["nsew", "xaf10000", "yaf10000"],
hist=True,
hist_bin_num=50,
)
# add subplot label
fig.text(
position="TL",
justify="TL",
text="b)",
font="16p,Helvetica,black",
offset="j.2c",
no_clip=True,
fill="white",
clearance="+tO",
pen=True,
)
####
# Starting bathymetry
####
fig = maps.plot_grd(
starting_bathymetry_no_errors,
fig=fig,
origin_shift="x",
region=plot_region,
title="Starting bathymetry",
cbar_label="elevation (m)",
cbar_font="18p,Helvetica,black",
cmap="rain",
reverse_cpt=True,
cpt_lims=bathy_lims,
frame=["nsew", "xaf10000", "yaf10000"],
hist=True,
points=constraint_points_dfs[0][~constraint_points_dfs[0].inside],
points_style="p2p",
)
fig.plot(
x=constraint_points_dfs[0][constraint_points_dfs[0].inside].easting,
y=constraint_points_dfs[0][constraint_points_dfs[0].inside].northing,
style="p4p",
pen="1p,black",
)
# add subplot label
fig.text(
position="TL",
justify="TL",
text="c)",
font="16p,Helvetica,black",
offset="j.2c",
no_clip=True,
fill="white",
clearance="+tO",
pen=True,
)
####
# Min distance
####
fig = maps.plot_grd(
min_dist,
fig=fig,
origin_shift="both",
xshift_amount=-1.7,
region=plot_region,
title="IBCSO points",
cbar_label="point proximity (m)",
cbar_font="18p,Helvetica,black",
cmap="dense",
frame=["nSWe", "xaf10000", "yaf10000"],
hist=True,
hist_bin_num=50,
cpt_lims=[
0,
polar_utils.get_min_max(
min_dist,
robust=True,
)[1],
],
points=total_ibcso_points,
points_style="p",
points_fill="black",
)
# add subplot label
fig.text(
position="TL",
justify="TL",
text="d)",
font="16p,Helvetica,black",
offset="j.2c",
no_clip=True,
fill="white",
clearance="+tO",
pen=True,
)
####
# Basement
####
fig = maps.plot_grd(
basement,
fig=fig,
origin_shift="x",
cmap="rain",
reverse_cpt=True,
cpt_lims=polar_utils.get_min_max(
basement, robust=True, robust_percentiles=(0.01, 0.99)
),
title="Basement",
cbar_label="elevation (m)",
cbar_font="18p,Helvetica,black",
frame=["nsew", "xaf10000", "yaf10000"],
hist=True,
inset=True,
inset_position="jTR+jTL",
inset_width=0.65,
)
# add subplot label
fig.text(
position="TL",
justify="TL",
text="e)",
font="16p,Helvetica,black",
offset="j.2c",
no_clip=True,
fill="white",
clearance="+tO",
pen=True,
)
fig.savefig("../paper/figures/Ross_Sea_location_figure.png", dpi=500)
fig.show()
Plotting functions¶
[11]:
profile_start = [plot_region[0], plot_region[3]]
profile_end = [plot_region[1], plot_region[2]]
[12]:
def sensitivity_plot(
titles,
grids,
constraint_points=None,
fname=None,
num_in_top_row=2,
plot_profile=False,
robust=True,
robust_percentiles=(0.02, 0.98),
):
grids = [
g.sel(
easting=slice(plot_region[0], plot_region[1]),
northing=slice(plot_region[2], plot_region[3]),
)
for g in grids
]
cpt_lims = polar_utils.get_combined_min_max(
grids[1],
robust=robust,
robust_percentiles=robust_percentiles,
)
fig_height = 9
for i, g in enumerate(grids):
xshift_amount = 1
if i == 0:
fig = None
origin_shift = "initialize"
scalebar = True
elif i == num_in_top_row:
origin_shift = "both_shift"
xshift_amount = -((len(grids) - 2) / 2)
scalebar = False
else:
origin_shift = "xshift"
scalebar = False
fig = maps.plot_grd(
grid=g,
fig_height=fig_height,
title=titles[i],
title_font="16p,Helvetica-bold,black",
cmap="ice",
reverse_cpt=True,
cpt_lims=(0, cpt_lims[1]),
cbar_unit="m",
cbar_font="18p,Helvetica,black",
hist=True,
hist_bin_num=50,
fig=fig,
origin_shift=origin_shift,
xshift_amount=xshift_amount,
yshift_amount=-1,
scalebar=scalebar,
# scalebar_position="jBR+o-.8c/-1.1c",
scalebar_position="jBR+o-1.3c/-1.1c",
scalebar_box=False,
points=constraint_points[~constraint_points.inside],
points_style="p1p",
)
fig.plot(
x=constraint_points[constraint_points.inside].easting,
y=constraint_points[constraint_points.inside].northing,
style="c.15c",
fill="white",
pen="1p,black",
)
# add mean / rms to plot
fig.text(
position="BR",
justify="TR",
text=f"RMS: {round(utils.rmse(g), 1)}",
fill="white",
font="16p,Helvetica,black",
offset="j0c/.2",
clearance="+tO",
no_clip=True,
)
if i == 0 and plot_profile:
# plot profiles location, and endpoints on map
fig.plot(
vd.line_coordinates(profile_start, profile_end, size=100),
pen="2p,black",
)
fig.text(
x=profile_start[0],
y=profile_start[1],
text="A",
fill="white",
font="12p,Helvetica,black",
justify="CM",
clearance="+tO",
no_clip=True,
)
fig.text(
x=profile_end[0],
y=profile_end[1],
text="A' ",
fill="white",
font="12p,Helvetica,black",
justify="CM",
clearance="+tO",
no_clip=True,
)
fig.text(
position="TL",
justify="TL",
text=f"{string.ascii_lowercase[i]}",
font="16p,Helvetica,black",
offset="j.2c",
no_clip=True,
fill="white",
clearance="+tO",
pen=True,
)
if fname is not None:
fig.savefig(fname)
return fig
[17]:
def sensitivity_profile(
titles,
grids,
constraint_points=None,
fname=None,
data_legend_loc="jTR+jTL+o1.5c/0c",
**kwargs,
):
grids = [
g.sel(
easting=slice(plot_region[0], plot_region[1]),
northing=slice(plot_region[2], plot_region[3]),
)
for g in grids
]
data_dict = profiles.make_data_dict(
names=titles,
grids=grids,
colors=[ # https://www.simplifiedsciencepublishing.com/resources/best-color-palettes-for-scientific-figures-and-data-visualizations
"216/48/52", # red, d83034
"0/58/125", # dark blue, 003a7d
"78/203/141", # green, 4ecb8d
"199/1/255", # purple, c701ff
"255/157/58", # orange, ff9d3a
"0/141/255", # med blue, 008dff
"255/115/182", # pink, ff73b6
"249/232/88", # yellow, f9e858
],
)
fig, df_data = profiles.plot_data(
"points",
start=profile_start,
stop=profile_end,
num=10000,
fig_height=4,
fig_width=14,
data_dict=data_dict,
data_legend_loc=data_legend_loc,
data_buffer=0.01,
data_frame=["neSW", "xafg+lDistance (m)", "yag+luncertainty (m)"],
data_pen_thickness=[2, 1.9, 1.2, 1.2, 1.2, 1.2, 1.2, 1.2],
share_yaxis=True,
start_label="A",
end_label="A' ",
**kwargs,
)
if constraint_points is not None:
# get grid of min dists to nearest constraints
min_dist = utils.normalized_mindist(
constraint_points,
grids[0],
)
# set y height in data units for add dist to constraint points
data_max = df_data.drop(columns=["x", "y", "dist"]).max().max()
if kwargs.get("data_ylims") is not None:
data_max = kwargs["data_ylims"][1]
df_data = profiles.sample_grids(
df_data, min_dist, interpolation="b", sampled_name="min_dist"
)
# set color limits
series = [1e3, 10e3]
pygmt.makecpt(
cmap="gray",
series=series,
background=True,
)
fig.shift_origin(
yshift=".2c",
)
fig.plot(
x=df_data.dist,
y=np.ones_like(df_data.dist) * data_max,
style="c0.1c",
fill=df_data.min_dist,
cmap=True,
no_clip=True,
)
fig.shift_origin(
yshift="-.2c",
)
with pygmt.config(
FONT="26p,Helvetica,black",
):
fig.colorbar(
cmap=True,
position="jMR+w2c/.3c+jML+v+o0.2c/0c+e",
frame=["xa+lnearest constraint", "y+lkm"],
scale=0.001,
)
if fname is not None:
fig.savefig(fname)
fig.show()
1: with density CV¶
[18]:
i = 1
final_bathymetry = final_bathymetries[i - 1]
inversion_results = optimal_results[i - 1]
damping_study = damping_studies[i - 1]
density_study = density_studies[i - 1]
constraint_points = pd.read_csv(
f"../results/Ross_Sea/Ross_Sea_0{i}_constraint_points.csv"
)
uncertainty = final_uncertainties[i - 1]
stochastic_mean = final_stochastic_means[i - 1]
sensitivity_ds = sensitivity_results[i - 1]
[ ]:
titles = [
"True error",
"Total uncertainty",
"Uncert. from damping",
"Uncert. from density",
]
grids = [
np.abs(stochastic_mean - bathymetry),
sensitivity_ds.full_stdev,
sensitivity_ds.damping_stdev,
sensitivity_ds.density_stdev,
]
sensitivity_plot(
titles,
grids,
constraint_points=constraint_points,
num_in_top_row=4,
# fname=f"../paper/figures/Ross_Sea_0{i}_sensitivity.png",
)
gmtset [WARNING]: Representation of font type not recognized. Using default.
gmtset [WARNING]: Representation of font type not recognized. Using default.
gmtset [WARNING]: Representation of font type not recognized. Using default.
gmtset [WARNING]: Representation of font type not recognized. Using default.
2: with noise¶
[20]:
i = 2
final_bathymetry = final_bathymetries[i - 1]
inversion_results = optimal_results[i - 1]
damping_study = damping_studies[i - 1]
density_study = density_studies[i - 1]
constraint_points = pd.read_csv(
f"../results/Ross_Sea/Ross_Sea_0{i}_constraint_points.csv"
)
uncertainty = final_uncertainties[i - 1]
stochastic_mean = final_stochastic_means[i - 1]
sensitivity_ds = sensitivity_results[i - 1]
[21]:
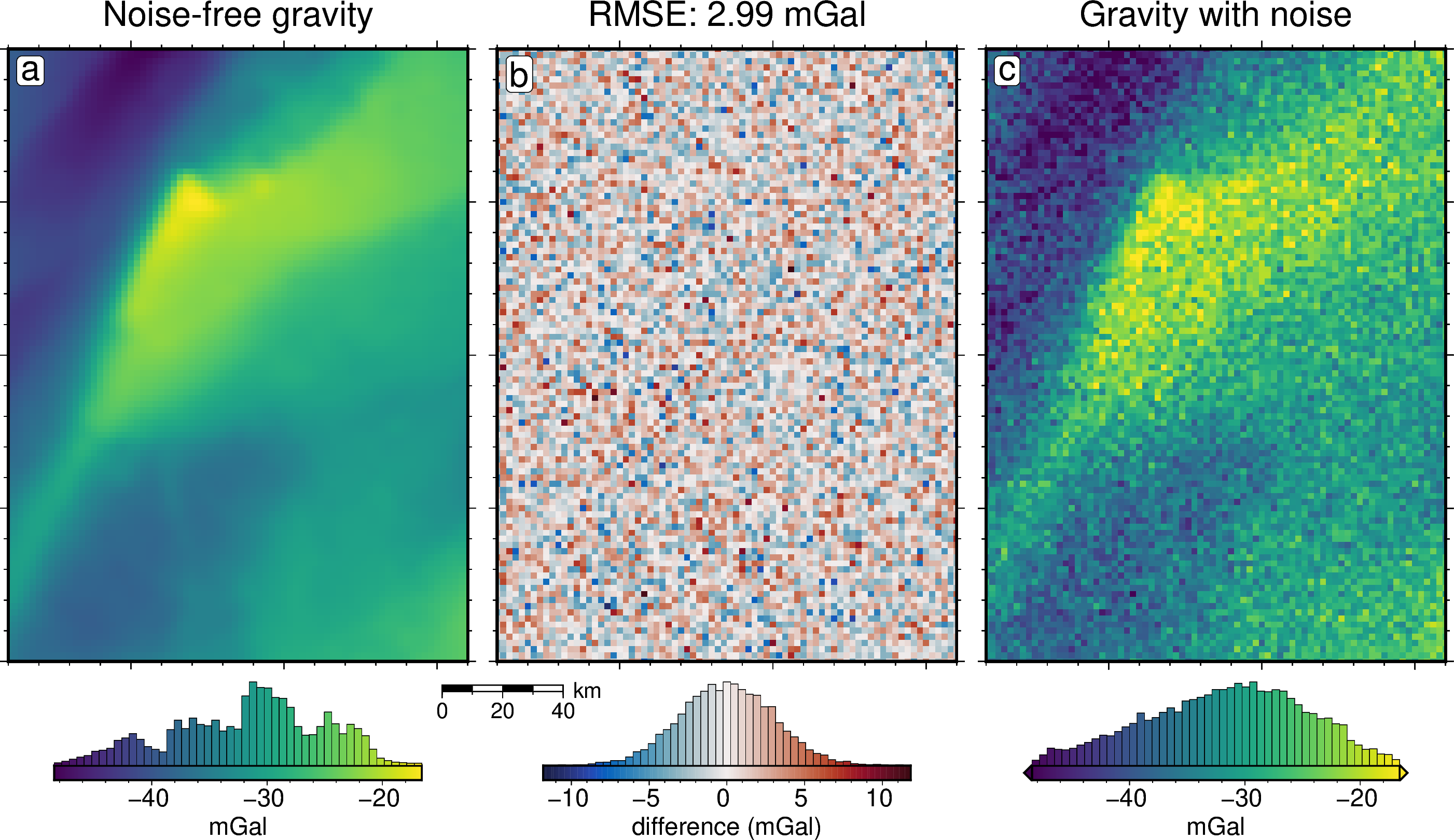
grav_df = inversion_results[1]
grav_grid = grav_df.set_index(["northing", "easting"]).to_xarray()
lims = polar_utils.get_min_max(grav_grid.noise_free_disturbance)
# plot the noise-free gravity
fig = maps.plot_grd(
grav_grid.noise_free_disturbance,
fig_height=10,
cpt_lims=lims,
title="Noise-free gravity",
title_font="16p,Helvetica,black",
cbar_label="mGal",
cbar_font="18p,Helvetica,black",
frame=["nswe", "xaf10000", "yaf10000"],
hist=True,
hist_bin_num=50,
)
fig.text(
position="TL",
text="a",
fill="white",
pen=True,
font="16p,Helvetica,black",
offset="j.2/.2",
clearance="+tO",
no_clip=True,
)
dif = grav_grid.noise_free_disturbance - grav_grid.disturbance
fig = maps.plot_grd(
dif,
fig_height=10,
cmap="balance+h0",
cpt_lims=polar_utils.get_min_max(
dif,
absolute=True,
),
title=f"RMSE: {round(utils.rmse(dif), 2)} mGal",
title_font="16p,Helvetica,black",
cbar_label="difference (mGal)",
cbar_font="18p,Helvetica,black",
frame=["nswe", "xaf10000", "yaf10000"],
hist=True,
hist_bin_num=50,
fig=fig,
origin_shift="xshift",
xshift_amount=1.01,
scalebar=True,
scalebar_position="jBL+o-.9c/-.6c",
)
fig.text(
position="TL",
text="b",
fill="white",
pen=True,
font="16p,Helvetica,black",
offset="j.2/.2",
clearance="+tO",
no_clip=True,
)
# plot the noisey gravity
fig = maps.plot_grd(
grav_grid.disturbance,
fig_height=10,
cpt_lims=lims,
title="Gravity with noise",
title_font="16p,Helvetica,black",
cbar_label="mGal",
cbar_font="18p,Helvetica,black",
frame=["nswe", "xaf10000", "yaf10000"],
hist=True,
hist_bin_num=50,
fig=fig,
origin_shift="xshift",
xshift_amount=1.01,
)
fig.text(
position="TL",
text="c",
fill="white",
pen=True,
font="16p,Helvetica,black",
offset="j.2/.2",
clearance="+tO",
no_clip=True,
)
fig.show()
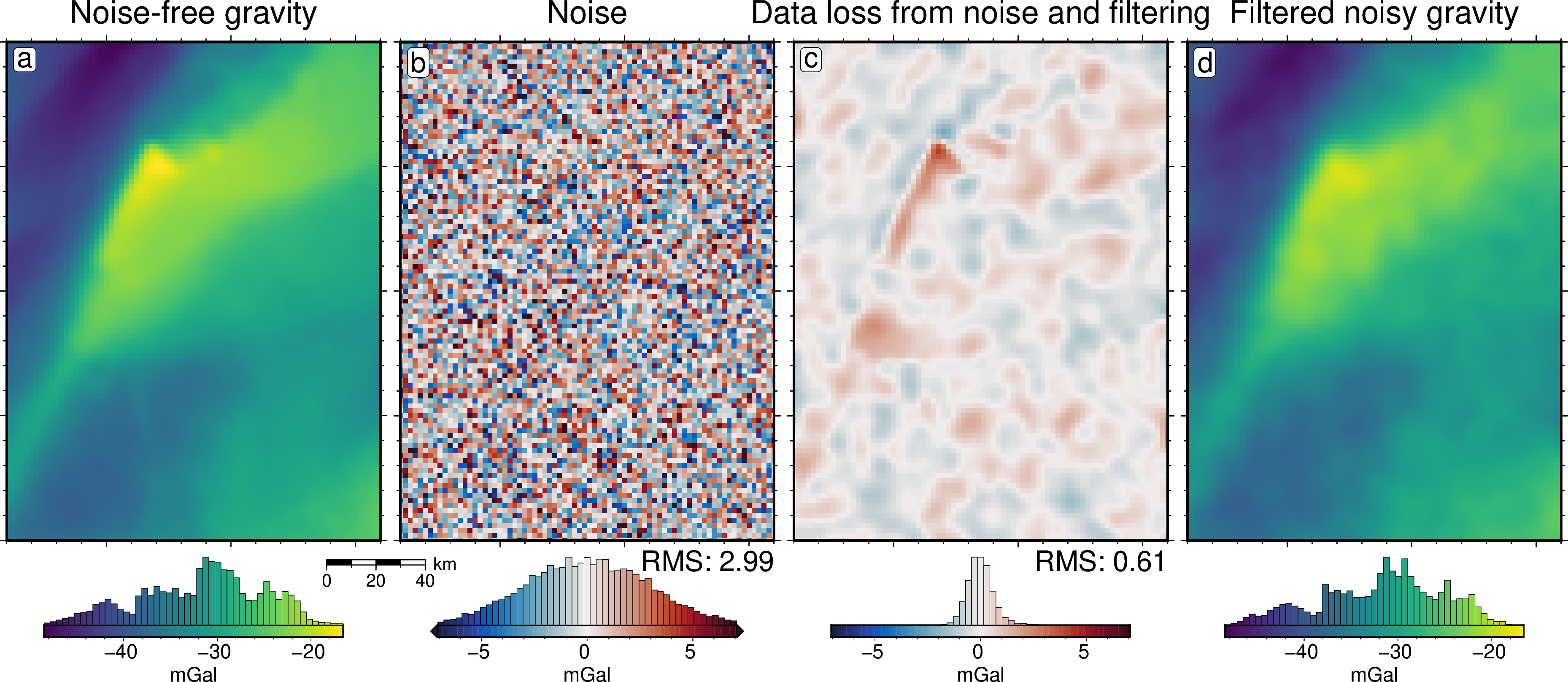
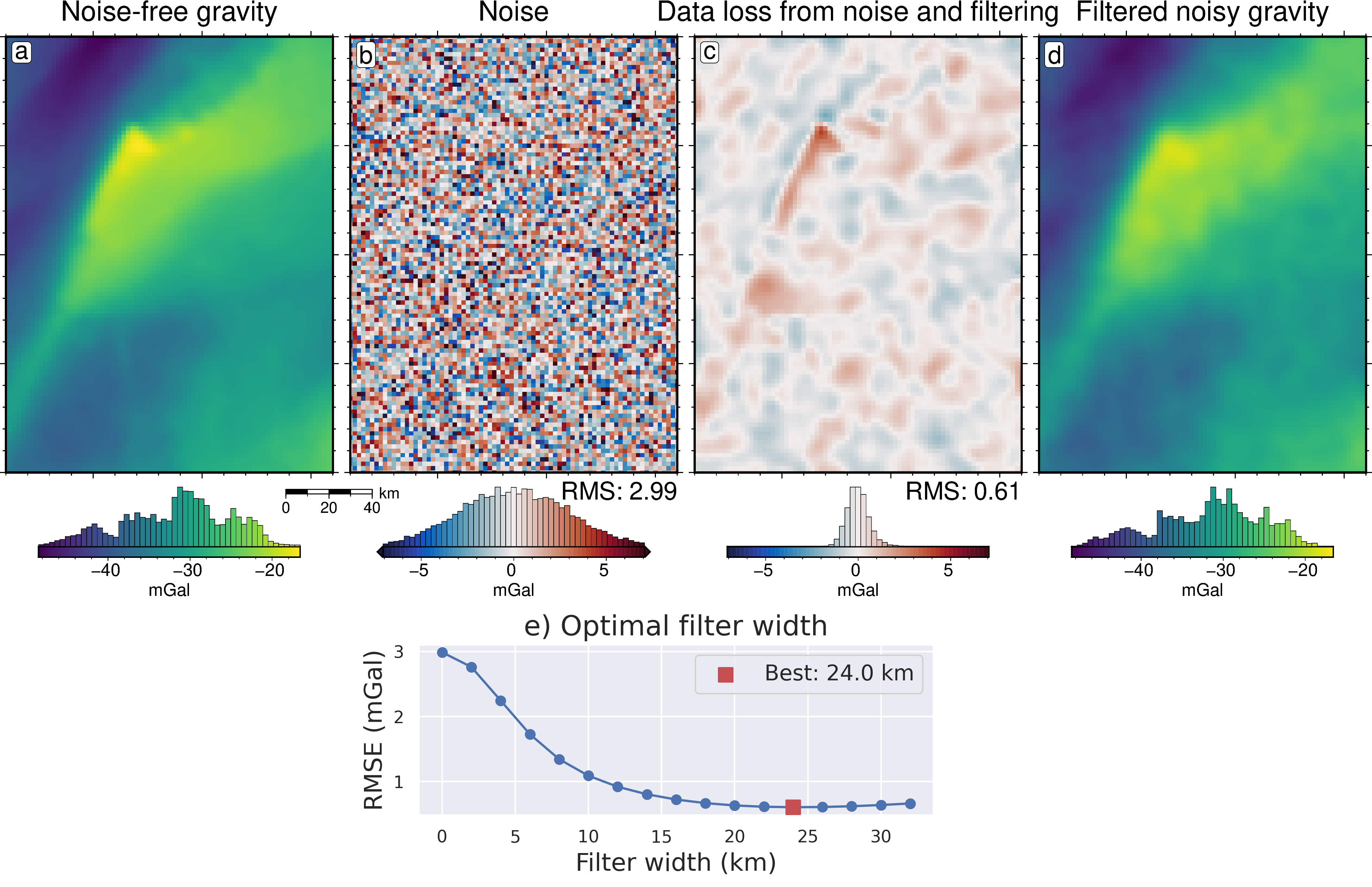
[22]:
grav_df = inversion_results[1]
grav_grid = grav_df.set_index(["northing", "easting"]).to_xarray()
lims = polar_utils.get_min_max(grav_grid.noise_free_disturbance)
# plot the noise-free gravity
fig = maps.plot_grd(
grav_grid.noise_free_disturbance,
fig_height=10,
cpt_lims=lims,
title="Noise-free gravity",
title_font="18p,Helvetica,black",
cbar_label="mGal",
cbar_font="18p,Helvetica,black",
frame=["nswe", "xaf10000", "yaf10000"],
hist=True,
scalebar=True,
scalebar_position="jBR+o-.9c/-.6c",
)
fig.text(
position="TL",
text="a",
fill="white",
pen=True,
font="16p,Helvetica,black",
offset="j.2/.2",
clearance="+tO",
no_clip=True,
)
# plot the difference between noise-free and noisy gravity
dif = grav_grid.noise_free_disturbance - grav_grid.disturbance
cpt_lims = polar_utils.get_min_max(
dif,
absolute=True,
robust=True,
robust_percentiles=(0.01, 0.99),
)
fig = maps.plot_grd(
dif,
fig_height=10,
cmap="balance+h0",
cpt_lims=cpt_lims,
title="Noise",
title_font="18p,Helvetica,black",
cbar_label="mGal",
cbar_font="18p,Helvetica,black",
frame=["nswe", "xaf10000", "yaf10000"],
hist=True,
fig=fig,
origin_shift="x",
)
fig.text(
position="TL",
text="b",
fill="white",
pen=True,
font="16p,Helvetica,black",
offset="j.2/.2",
clearance="+tO",
no_clip=True,
)
# add mean / rms to plot
fig.text(
position="BR",
justify="TR",
text=f"RMS: {round(utils.rmse(dif), 2)}",
fill="white",
font="16p,Helvetica,black",
offset="j0c/.2",
clearance="+tO",
no_clip=True,
)
# plot the difference between noisy and filtered gravity
dif = grav_grid.noise_free_disturbance - grav_grid.gravity_anomaly
fig = maps.plot_grd(
dif,
fig_height=10,
cmap="balance+h0",
cpt_lims=cpt_lims,
title="Data loss from noise and filtering",
title_font="18p,Helvetica,black",
cbar_label="mGal",
cbar_font="18p,Helvetica,black",
frame=["nswe", "xaf10000", "yaf10000"],
hist=True,
fig=fig,
origin_shift="x",
)
fig.text(
position="TL",
text="c",
fill="white",
pen=True,
font="16p,Helvetica,black",
offset="j.2/.2",
clearance="+tO",
no_clip=True,
)
# add mean / rms to plot
fig.text(
position="BR",
justify="TR",
text=f"RMS: {round(utils.rmse(dif), 2)}",
fill="white",
font="16p,Helvetica,black",
offset="j0c/.2",
clearance="+tO",
no_clip=True,
)
# plot the filtered gravity
fig = maps.plot_grd(
grav_grid.gravity_anomaly,
fig_height=10,
cpt_lims=lims,
title="Filtered noisy gravity",
title_font="18p,Helvetica,black",
cbar_label="mGal",
cbar_font="18p,Helvetica,black",
frame=["nswe", "xaf10000", "yaf10000"],
hist=True,
fig=fig,
origin_shift="xshift",
)
fig.text(
position="TL",
text="d",
fill="white",
pen=True,
font="16p,Helvetica,black",
offset="j.2/.2",
clearance="+tO",
no_clip=True,
)
fig.show()
[ ]:
filter_widths_df = pd.read_csv(
f"../results/Ross_Sea/Ross_Sea_0{i}_filter_width_rmse.csv"
)
_fig, ax = plt.subplots(figsize=(6, 2))
ax.plot(
filter_widths_df.filt_width / 1e3,
filter_widths_df.rmse,
marker="o",
)
best_ind = filter_widths_df.rmse.idxmin()
ax.scatter(
x=filter_widths_df.iloc[best_ind].filt_width / 1e3,
y=filter_widths_df.iloc[best_ind].rmse,
marker="s",
color="r",
s=80,
zorder=20,
label=f"Best: {round(filter_widths_df.iloc[best_ind].filt_width / 1e3, 2)} km",
)
ax.legend(fontsize=14)
ax.set_xlabel("Filter width (km)", fontsize=16)
ax.set_ylabel("RMSE (mGal)", fontsize=16)
ax.set_title("e) Optimal filter width", fontsize=18)
_fig.savefig(
f"../paper/figures/Ross_Sea_0{i}_filters.png", dpi=300, bbox_inches="tight"
)
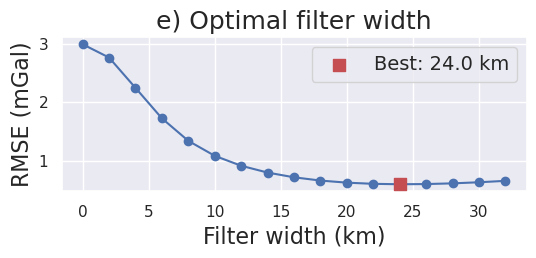
[24]:
fig.image(
imagefile=f"../paper/figures/Ross_Sea_0{i}_filters.png",
position="n-2.1/-.3+jTL",
)
fig.savefig(f"../paper/figures/Ross_Sea_0{i}_grav_error.png")
fig.show()
[25]:
grav_df = inversion_results[1]
noise = np.abs(grav_df.noise_free_disturbance - grav_df.disturbance)
noise.describe()
[25]:
count 7676.000000
mean 2.387429
std 1.801864
min 0.000615
25% 0.955809
50% 2.028027
75% 3.421094
max 11.818300
dtype: float64
[26]:
utils.rmse(noise)
[26]:
np.float64(2.99100483668976)
[27]:
grav_error = np.abs(grav_df.noise_free_disturbance - grav_df.gravity_anomaly)
grav_error.describe()
[27]:
count 7676.000000
mean 0.463315
std 0.389367
min 0.000173
25% 0.181996
50% 0.369205
75% 0.642503
max 4.125650
dtype: float64
[28]:
utils.rmse(grav_error)
[28]:
np.float64(0.6051841170730238)
[ ]:
titles = [
"True error",
"Total uncertainty",
"Uncert. from damping",
"Uncert. from density",
"Uncert. from gravity",
]
grids = [
np.abs(stochastic_mean - bathymetry),
sensitivity_ds.full_stdev,
sensitivity_ds.damping_stdev,
sensitivity_ds.density_stdev,
sensitivity_ds.gravity_stdev,
]
sensitivity_plot(
titles,
grids,
constraint_points=constraint_points,
num_in_top_row=2,
# fname=f"../paper/figures/Ross_Sea_0{i}_sensitivity.png",
)
gmtset [WARNING]: Representation of font type not recognized. Using default.
gmtset [WARNING]: Representation of font type not recognized. Using default.
gmtset [WARNING]: Representation of font type not recognized. Using default.
gmtset [WARNING]: Representation of font type not recognized. Using default.
gmtset [WARNING]: Representation of font type not recognized. Using default.
3: with airborne survey¶
[30]:
i = 3
final_bathymetry = final_bathymetries[i - 1]
inversion_results = optimal_results[i - 1]
damping_study = damping_studies[i - 1]
density_study = density_studies[i - 1]
with pathlib.Path(f"../results/Ross_Sea/Ross_Sea_0{i}_eq_sources.pickle").open(
"rb"
) as f:
eqs_study = pickle.load(f)
constraint_points = pd.read_csv(
f"../results/Ross_Sea/Ross_Sea_0{i}_constraint_points.csv"
)
uncertainty = final_uncertainties[i - 1]
stochastic_mean = final_stochastic_means[i - 1]
grav_df = pd.read_csv(f"../results/Ross_Sea/Ross_Sea_0{i}_grav_df.csv")
grav_survey_df = pd.read_csv(f"../results/Ross_Sea/Ross_Sea_0{i}_grav_survey_df.csv")
sensitivity_ds = sensitivity_results[i - 1]
[31]:
grav_df.describe()
[31]:
| northing | easting | upward | bathymetry_grav | basement_grav | disturbance | gravity_anomaly_full_res | eqs_gravity_anomaly | gravity_anomaly_deterministic | starting_gravity | misfit | reg | res | predicted_grav | eqs_uncert | gravity_anomaly_eqs_mean | gravity_anomaly | uncert | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| count | 7.676000e+03 | 7676.000000 | 7676.0 | 7676.000000 | 7676.0 | 7676.000000 | 7676.000000 | 7676.000000 | 7676.000000 | 7676.000000 | 7676.000000 | 7676.000000 | 7676.000000 | 7676.000000 | 7676.000000 | 7676.000000 | 7676.000000 | 7676.000000 |
| mean | -1.500000e+06 | 35000.000000 | 1000.0 | -31.727281 | 0.0 | -31.727281 | -31.727281 | -31.706558 | -31.706558 | -31.307367 | -0.393674 | -0.197494 | -0.196180 | -31.699659 | 0.053538 | -31.701041 | -31.701041 | 0.053538 |
| std | 5.831332e+04 | 43877.680138 | 0.0 | 6.458148 | 0.0 | 6.458148 | 6.458148 | 6.432334 | 6.432334 | 6.220031 | 1.237689 | 0.000000 | 1.237689 | 6.423451 | 0.076367 | 6.425508 | 6.425508 | 0.076367 |
| min | -1.600000e+06 | -40000.000000 | 1000.0 | -48.457772 | 0.0 | -48.457772 | -48.457772 | -48.094852 | -48.094852 | -48.209648 | -7.268971 | -0.197494 | -7.071478 | -48.134527 | 0.001516 | -48.085594 | -48.085594 | 0.001516 |
| 25% | -1.550000e+06 | -2500.000000 | 1000.0 | -36.286366 | 0.0 | -36.286366 | -36.286366 | -36.308653 | -36.308653 | -35.436143 | -0.755584 | -0.197494 | -0.558091 | -36.285323 | 0.018445 | -36.297954 | -36.297954 | 0.018445 |
| 50% | -1.500000e+06 | 35000.000000 | 1000.0 | -31.030069 | 0.0 | -31.030069 | -31.030069 | -30.978656 | -30.978656 | -30.827291 | -0.295344 | -0.197494 | -0.097850 | -30.980281 | 0.031697 | -30.977286 | -30.977286 | 0.031697 |
| 75% | -1.450000e+06 | 72500.000000 | 1000.0 | -27.278975 | 0.0 | -27.278975 | -27.278975 | -27.240553 | -27.240553 | -26.861446 | 0.038455 | -0.197494 | 0.235949 | -27.233120 | 0.056035 | -27.237610 | -27.237610 | 0.056035 |
| max | -1.400000e+06 | 110000.000000 | 1000.0 | -16.585798 | 0.0 | -16.585798 | -16.585798 | -16.551546 | -16.551546 | -16.852773 | 5.599752 | -0.197494 | 5.797246 | -16.787896 | 0.833729 | -16.734482 | -16.734482 | 0.833729 |
[32]:
grav_grid = grav_df.set_index(["northing", "easting"]).to_xarray()
lims = polar_utils.get_min_max(grav_grid.gravity_anomaly_full_res)
# plot the noise-free gravity
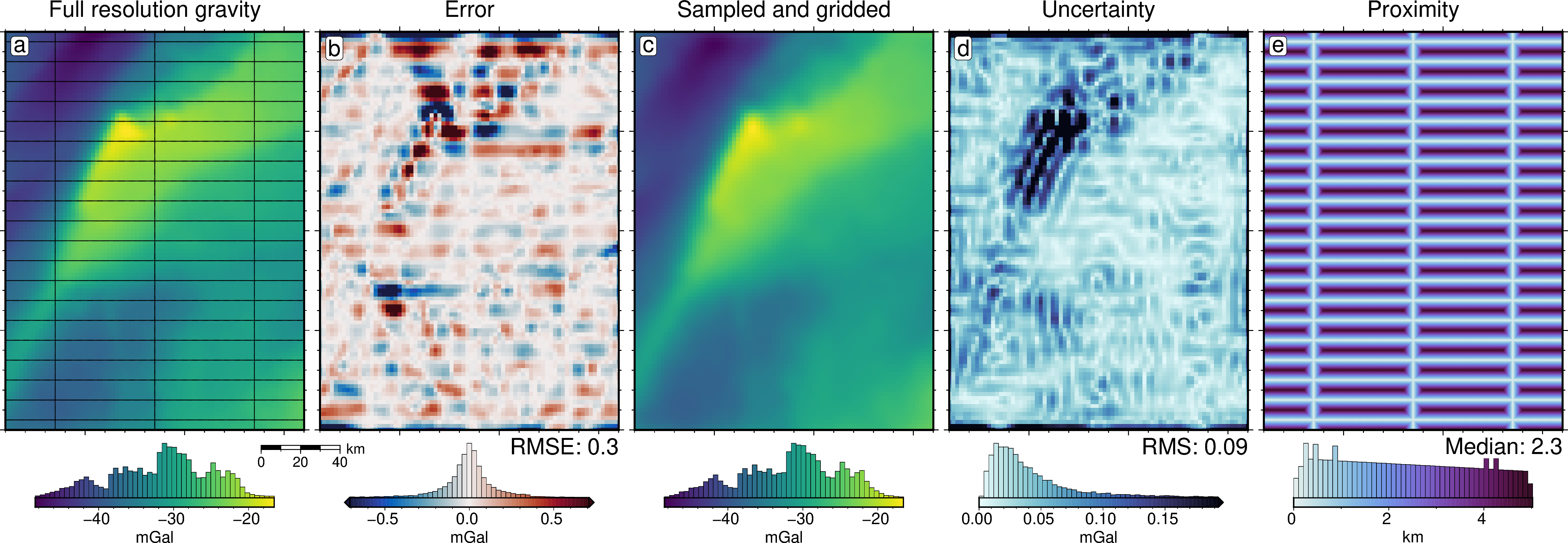
fig = maps.plot_grd(
grav_grid.gravity_anomaly_full_res,
fig_height=10,
cpt_lims=lims,
title="Full resolution gravity",
title_font="16p,Helvetica,black",
cbar_label="mGal",
cbar_font="18p,Helvetica,black",
frame=["nswe", "xaf10000", "yaf10000"],
hist=True,
scalebar=True,
scalebar_position="jBR+o-.9c/-.6c",
)
# plot observation points
fig.plot(
grav_survey_df[["easting", "northing"]],
style="c.02c",
fill="black",
)
fig.text(
position="TL",
text="a",
fill="white",
pen=True,
font="16p,Helvetica,black",
offset="j.2/.2",
clearance="+tO",
no_clip=True,
)
dif = grav_grid.gravity_anomaly_full_res - grav_grid.gravity_anomaly
dif_lims = polar_utils.get_min_max(
dif,
absolute=True,
robust=True,
)
fig = maps.plot_grd(
dif,
fig_height=10,
cmap="balance+h0",
cpt_lims=dif_lims,
title="Error", # , RMSE: {round(utils.rmse(dif),1)} mGal",
title_font="16p,Helvetica,black",
cbar_label="mGal",
cbar_font="18p,Helvetica,black",
frame=["nswe", "xaf10000", "yaf10000"],
hist=True,
fig=fig,
origin_shift="x",
)
# add mean / rms to plot
fig.text(
position="BR",
justify="TR",
text=f"RMSE: {round(utils.rmse(dif), 2)}",
fill="white",
font="16p,Helvetica,black",
offset="j0c/.2",
clearance="+tO",
no_clip=True,
)
fig.text(
position="TL",
text="b",
fill="white",
pen=True,
font="16p,Helvetica,black",
offset="j.2/.2",
clearance="+tO",
no_clip=True,
)
# plot the noisey gravity
fig = maps.plot_grd(
grav_grid.gravity_anomaly,
fig_height=10,
cpt_lims=lims,
title="Sampled and gridded",
title_font="16p,Helvetica,black",
cbar_label="mGal",
cbar_font="18p,Helvetica,black",
frame=["nswe", "xaf10000", "yaf10000"],
hist=True,
fig=fig,
origin_shift="x",
)
fig.text(
position="TL",
text="c",
fill="white",
pen=True,
font="16p,Helvetica,black",
offset="j.2/.2",
clearance="+tO",
no_clip=True,
)
# plot the eq source uncertainty
fig = maps.plot_grd(
grav_grid.uncert,
fig_height=10,
cpt_lims=(
0,
polar_utils.get_min_max(
grav_grid.uncert,
robust=True,
robust_percentiles=(0.04, 0.96),
)[1],
),
cmap="ice",
reverse_cpt=True,
title="Uncertainty", # , RMS: {round(utils.rmse(grav_grid.uncert),2)} mGal",
title_font="16p,Helvetica,black",
cbar_label="mGal", # , RMS: {round(utils.rmse(grav_grid.uncert),2)}",
cbar_font="18p,Helvetica,black",
frame=["nswe", "xaf10000", "yaf10000"],
hist=True,
fig=fig,
origin_shift="x",
)
# add mean / rms to plot
fig.text(
position="BR",
justify="TR",
text=f"RMS: {round(utils.rmse(grav_grid.uncert), 2)}",
fill="white",
font="16p,Helvetica,black",
offset="j0c/.2",
clearance="+tO",
no_clip=True,
)
fig.text(
position="TL",
text="d",
fill="white",
pen=True,
font="16p,Helvetica,black",
offset="j.2/.2",
clearance="+tO",
no_clip=True,
)
coords = vd.grid_coordinates(
region=inversion_region,
spacing=100,
)
grid = vd.make_xarray_grid(coords, np.ones_like(coords[0]), data_names="z").z
min_dist = (
utils.dist_nearest_points(
grav_survey_df,
grid,
).min_dist
/ 1e3
)
fig = maps.plot_grd(
min_dist,
fig_height=10,
cpt_lims=(
0,
polar_utils.get_min_max(
min_dist,
)[1],
),
cmap="dense",
title="Proximity", # , median: {round(np.median(min_dist),2)} km",
title_font="16p,Helvetica,black",
cbar_label="km",
cbar_font="18p,Helvetica,black",
frame=["nswe", "xaf10000", "yaf10000"],
hist=True,
fig=fig,
origin_shift="x",
)
# add mean / rms to plot
fig.text(
position="BR",
justify="TR",
text=f"Median: {round(np.median(min_dist), 2)}",
fill="white",
font="16p,Helvetica,black",
offset="j0c/.2",
clearance="+tO",
no_clip=True,
)
fig.text(
position="TL",
text="e",
fill="white",
pen=True,
font="16p,Helvetica,black",
offset="j.2/.2",
clearance="+tO",
no_clip=True,
)
fig.savefig(f"../paper/figures/Ross_Sea_0{i}_grav_error.png")
fig.show()
[33]:
df = eqs_study.trials_dataframe().sort_values("value").reset_index(drop=True)
df = df.iloc[2:].reset_index(drop=True)
df = df.drop(3).reset_index(drop=True)
df
[33]:
| number | value | datetime_start | datetime_complete | duration | params_damping | params_depth | system_attrs_fixed_params | state | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 7 | 0.975067 | 2025-10-14 14:30:30.659713 | 2025-10-14 14:30:33.377640 | 0 days 00:00:02.717927 | 0.098499 | 72439.765810 | NaN | COMPLETE |
| 1 | 3 | 0.976299 | 2025-10-14 14:30:01.380711 | 2025-10-14 14:30:06.333186 | 0 days 00:00:04.952475 | 0.762644 | 59952.265810 | NaN | COMPLETE |
| 2 | 5 | 0.976426 | 2025-10-14 14:30:12.405263 | 2025-10-14 14:30:21.988305 | 0 days 00:00:09.583042 | 0.004572 | 84927.265810 | NaN | COMPLETE |
| 3 | 4 | 0.994434 | 2025-10-14 14:30:06.340498 | 2025-10-14 14:30:12.401418 | 0 days 00:00:06.060920 | 0.035399 | 34977.265810 | NaN | COMPLETE |
| 4 | 6 | 0.995391 | 2025-10-14 14:30:22.019279 | 2025-10-14 14:30:30.658220 | 0 days 00:00:08.638941 | 2.122102 | 22489.765810 | NaN | COMPLETE |
| 5 | 9 | 0.999350 | 2025-10-14 14:30:40.258524 | 2025-10-14 14:30:48.611805 | 0 days 00:00:08.353281 | 0.411685 | 14173.067440 | NaN | COMPLETE |
| 6 | 8 | 0.999677 | 2025-10-14 14:30:33.405174 | 2025-10-14 14:30:40.256842 | 0 days 00:00:06.851668 | 0.001000 | 19115.478929 | NaN | COMPLETE |
| 7 | 11 | 0.999952 | 2025-10-14 14:30:58.998486 | 2025-10-14 14:31:05.847557 | 0 days 00:00:06.849071 | 0.001000 | 13499.630288 | NaN | COMPLETE |
| 8 | 2 | 0.999986 | 2025-10-14 14:29:56.771327 | 2025-10-14 14:30:01.375794 | 0 days 00:00:04.604467 | 0.001643 | 10002.265810 | NaN | COMPLETE |
[34]:
spline = vd.Spline(damping=None)
coords = (
df.params_depth,
df.params_damping,
)
spline.fit(
coords,
df.value,
)
grid_coords = vd.grid_coordinates(
region=vd.get_region(coords),
shape=(100, 100),
pixel_register=True,
)
grid = vd.make_xarray_grid(
grid_coords,
data=np.ones_like(grid_coords[0]),
data_names="data",
dims=("damping", "depth"), # these need to be swapped
).data
score_df = vd.grid_to_table(grid).drop(columns="data")
score_df["score"] = spline.predict((score_df.damping, score_df.depth))
grid = score_df.set_index(["damping", "depth"]).to_xarray().score
[35]:
df
[35]:
| number | value | datetime_start | datetime_complete | duration | params_damping | params_depth | system_attrs_fixed_params | state | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 7 | 0.975067 | 2025-10-14 14:30:30.659713 | 2025-10-14 14:30:33.377640 | 0 days 00:00:02.717927 | 0.098499 | 72439.765810 | NaN | COMPLETE |
| 1 | 3 | 0.976299 | 2025-10-14 14:30:01.380711 | 2025-10-14 14:30:06.333186 | 0 days 00:00:04.952475 | 0.762644 | 59952.265810 | NaN | COMPLETE |
| 2 | 5 | 0.976426 | 2025-10-14 14:30:12.405263 | 2025-10-14 14:30:21.988305 | 0 days 00:00:09.583042 | 0.004572 | 84927.265810 | NaN | COMPLETE |
| 3 | 4 | 0.994434 | 2025-10-14 14:30:06.340498 | 2025-10-14 14:30:12.401418 | 0 days 00:00:06.060920 | 0.035399 | 34977.265810 | NaN | COMPLETE |
| 4 | 6 | 0.995391 | 2025-10-14 14:30:22.019279 | 2025-10-14 14:30:30.658220 | 0 days 00:00:08.638941 | 2.122102 | 22489.765810 | NaN | COMPLETE |
| 5 | 9 | 0.999350 | 2025-10-14 14:30:40.258524 | 2025-10-14 14:30:48.611805 | 0 days 00:00:08.353281 | 0.411685 | 14173.067440 | NaN | COMPLETE |
| 6 | 8 | 0.999677 | 2025-10-14 14:30:33.405174 | 2025-10-14 14:30:40.256842 | 0 days 00:00:06.851668 | 0.001000 | 19115.478929 | NaN | COMPLETE |
| 7 | 11 | 0.999952 | 2025-10-14 14:30:58.998486 | 2025-10-14 14:31:05.847557 | 0 days 00:00:06.849071 | 0.001000 | 13499.630288 | NaN | COMPLETE |
| 8 | 2 | 0.999986 | 2025-10-14 14:29:56.771327 | 2025-10-14 14:30:01.375794 | 0 days 00:00:04.604467 | 0.001643 | 10002.265810 | NaN | COMPLETE |
[36]:
_fig, ax = plt.subplots(figsize=(3.5, 5))
grid.plot(
cmap=sns.color_palette("mako", as_cmap=True),
ax=ax,
)
best_ind = df.value.idxmax()
ax.scatter(
y=df.iloc[best_ind].params_damping,
x=df.iloc[best_ind].params_depth,
marker="s",
s=100,
color="r",
label="Best",
)
ax.scatter(
y=df.params_damping,
x=df.params_depth,
marker=".",
color="lightgray",
edgecolor="black",
)
depth_range = df.params_depth.max() - df.params_depth.min()
ax.set_xlim(
df.params_depth.min() - (depth_range * 0.02),
df.params_depth.max() + (depth_range * 0.02),
)
damping_range = df.params_damping.max() - df.params_damping.min()
ax.set_ylim(
df.params_damping.min() - (damping_range * 0.02),
df.params_damping.max() + (damping_range * 0.02),
)
[36]:
(-0.04142203060414228, 2.164523560811256)
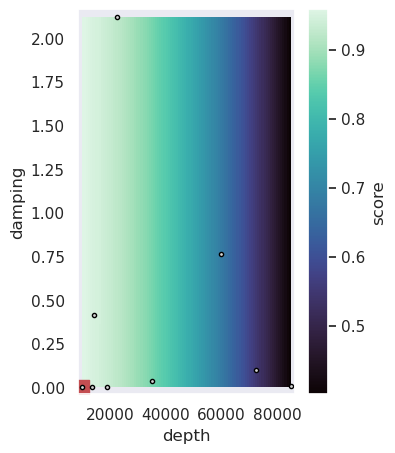
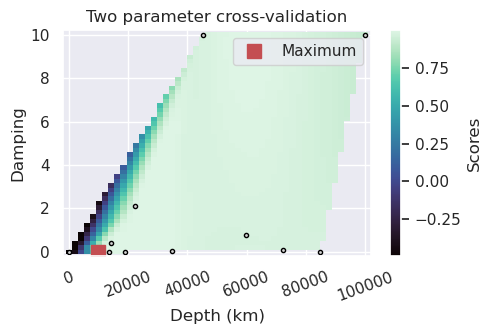
[37]:
plotting.plot_2_parameter_cv_scores_uneven(
eqs_study,
param_names=(
"params_depth",
"params_damping",
),
plot_param_names=(
"Depth (km)",
"Damping",
),
best="max",
robust=True,
)
[38]:
grav_error = np.abs(grav_df.gravity_anomaly_full_res - grav_df.gravity_anomaly)
print(utils.rmse(grav_error))
grav_error.describe()
0.29555401447894813
[38]:
count 7676.000000
mean 0.140733
std 0.259914
min 0.000016
25% 0.027358
50% 0.066101
75% 0.151545
max 4.329160
dtype: float64
[ ]:
titles = [
"True error",
"Total uncertainty",
"Uncert. from damping",
"Uncert. from density",
"Uncert. from interpolation",
]
grids = [
np.abs(stochastic_mean - bathymetry),
sensitivity_ds.full_stdev,
sensitivity_ds.damping_stdev,
sensitivity_ds.density_stdev,
sensitivity_ds["equivalent source gridding_stdev"],
]
sensitivity_plot(
titles,
grids,
constraint_points=constraint_points,
num_in_top_row=2,
# fname=f"../paper/figures/Ross_Sea_0{i}_sensitivity.png",
)
gmtset [WARNING]: Representation of font type not recognized. Using default.
gmtset [WARNING]: Representation of font type not recognized. Using default.
gmtset [WARNING]: Representation of font type not recognized. Using default.
gmtset [WARNING]: Representation of font type not recognized. Using default.
gmtset [WARNING]: Representation of font type not recognized. Using default.
4: with regional¶
[18]:
i = 4
final_bathymetry = final_bathymetries[i - 1]
inversion_results = optimal_results[i - 1]
damping_study = damping_studies[i - 1]
density_study = density_studies[i - 1]
with pathlib.Path(
f"../tmp_outputs/Ross_Sea_0{i}_regional_sep_true_density.pickle"
).open("rb") as f:
regional_study = pickle.load(f)
constraint_points = pd.read_csv(
f"../results/Ross_Sea/Ross_Sea_0{i}_constraint_points.csv"
)
uncertainty = final_uncertainties[i - 1]
stochastic_mean = final_stochastic_means[i - 1]
grav_df = pd.read_csv(f"../results/Ross_Sea/Ross_Sea_0{i}_grav_df_true_density.csv")
sensitivity_ds = sensitivity_results[i - 1]
[19]:
grav_df.head()
[19]:
| northing | easting | upward | bathymetry_grav | basement_grav_normalized | basement_grav | gravity_anomaly | starting_gravity | true_res | misfit | reg | res | reg_uncert | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | -1600000.0 | -40000.0 | 1000.0 | -35.551054 | -0.575610 | -36.298772 | -71.849826 | -35.578561 | -3.009687 | -36.271265 | -36.352514 | 0.081249 | 0.196374 |
| 1 | -1600000.0 | -38000.0 | 1000.0 | -36.054657 | -0.523194 | -33.940048 | -69.994704 | -36.042642 | -3.088826 | -33.952063 | -33.935629 | -0.016434 | 0.082311 |
| 2 | -1600000.0 | -36000.0 | 1000.0 | -36.473146 | -0.466341 | -31.381660 | -67.854806 | -36.460095 | -3.125498 | -31.394711 | -31.343652 | -0.051059 | 0.058318 |
| 3 | -1600000.0 | -34000.0 | 1000.0 | -36.755608 | -0.406001 | -28.666395 | -65.422003 | -36.784649 | -3.111113 | -28.637354 | -28.615142 | -0.022212 | 0.076759 |
| 4 | -1600000.0 | -32000.0 | 1000.0 | -36.951029 | -0.343146 | -25.837899 | -62.788928 | -36.998469 | -3.110966 | -25.790460 | -25.788302 | -0.002158 | 0.084586 |
[20]:
# calculate the true residual and regional misfit
grav_df["true_res"] = grav_df.bathymetry_grav - grav_df.starting_gravity
grav_df["true_reg"] = grav_df.basement_grav
grav_df.head()
[20]:
| northing | easting | upward | bathymetry_grav | basement_grav_normalized | basement_grav | gravity_anomaly | starting_gravity | true_res | misfit | reg | res | reg_uncert | true_reg | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | -1600000.0 | -40000.0 | 1000.0 | -35.551054 | -0.575610 | -36.298772 | -71.849826 | -35.578561 | 0.027507 | -36.271265 | -36.352514 | 0.081249 | 0.196374 | -36.298772 |
| 1 | -1600000.0 | -38000.0 | 1000.0 | -36.054657 | -0.523194 | -33.940048 | -69.994704 | -36.042642 | -0.012015 | -33.952063 | -33.935629 | -0.016434 | 0.082311 | -33.940048 |
| 2 | -1600000.0 | -36000.0 | 1000.0 | -36.473146 | -0.466341 | -31.381660 | -67.854806 | -36.460095 | -0.013051 | -31.394711 | -31.343652 | -0.051059 | 0.058318 | -31.381660 |
| 3 | -1600000.0 | -34000.0 | 1000.0 | -36.755608 | -0.406001 | -28.666395 | -65.422003 | -36.784649 | 0.029040 | -28.637354 | -28.615142 | -0.022212 | 0.076759 | -28.666395 |
| 4 | -1600000.0 | -32000.0 | 1000.0 | -36.951029 | -0.343146 | -25.837899 | -62.788928 | -36.998469 | 0.047440 | -25.790460 | -25.788302 | -0.002158 | 0.084586 | -25.837899 |
[21]:
print(f"MA of regional: {np.abs(grav_df.reg).mean()}")
print(f"RMS of regional: {utils.rmse(grav_df.reg)}")
print(f"MAE of regional: {np.abs(grav_df.true_reg - grav_df.reg).mean()}")
print(f"RMSE of regional: {utils.rmse(grav_df.true_reg - grav_df.reg)}")
print(f"Max regional error: {np.abs(grav_df.true_reg - grav_df.reg).max()}")
MA of regional: 15.359499569436794
RMS of regional: 18.35363723599696
MAE of regional: 0.3368661422817604
RMSE of regional: 0.4687092556750809
Max regional error: 2.754957899309744
[22]:
grav_grid = grav_df.set_index(["northing", "easting"]).to_xarray()
grav_grid["true_reg"] = grav_grid.basement_grav
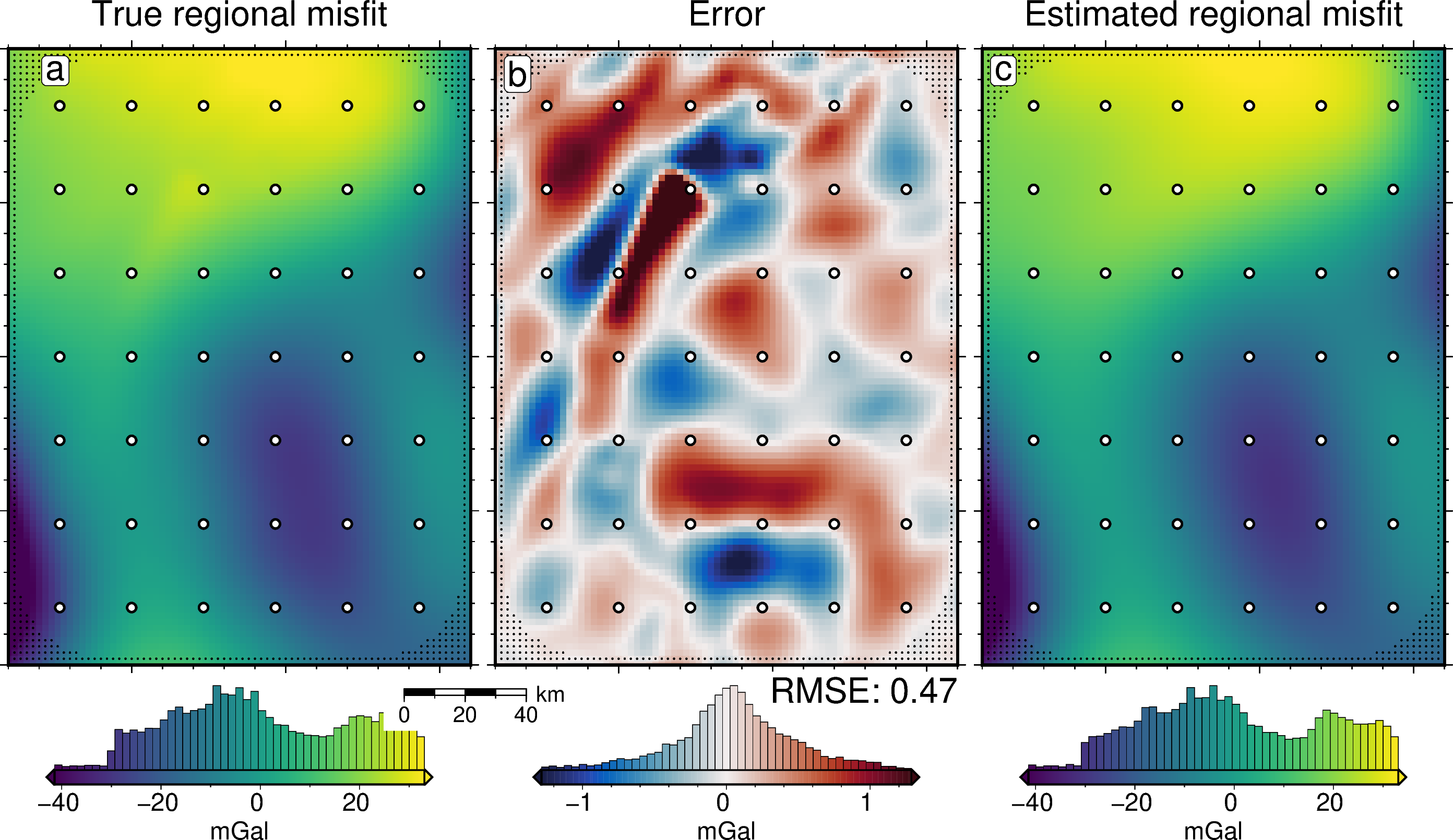
[14]:
reg_lims = polar_utils.get_combined_min_max(
[grav_grid.true_reg, grav_grid.reg],
robust=True,
robust_percentiles=(0.01, 0.99),
)
fig = maps.plot_grd(
grav_grid.true_reg,
fig_height=10,
cmap="viridis",
cpt_lims=reg_lims,
title="True regional misfit",
title_font="16p,Helvetica,black",
cbar_label="mGal",
cbar_font="18p,Helvetica,black",
frame=["nswe", "xaf10000", "yaf10000"],
hist=True,
points=constraint_points[~constraint_points.inside],
points_style="p1p",
scalebar=True,
scalebar_position="jBR+o-.9c/-.6c",
)
fig.plot(
x=constraint_points[constraint_points.inside].easting,
y=constraint_points[constraint_points.inside].northing,
style="c.15c",
fill="white",
pen="1p,black",
)
fig.text(
position="TL",
text="a",
fill="white",
pen=True,
font="16p,Helvetica,black",
offset="j.6/.2",
clearance="+tO",
no_clip=True,
)
# plot the regional error
dif = grav_grid.true_reg - grav_grid.reg
fig = maps.plot_grd(
dif,
fig_height=10,
cmap="balance+h0",
cpt_lims=polar_utils.get_min_max(
dif,
absolute=True,
robust=True,
robust_percentiles=(0.01, 0.99),
),
title="Error",
title_font="16p,Helvetica,black",
cbar_label="mGal", # f"mGal, RMSE: {round(utils.rmse(dif),2)}",
cbar_font="18p,Helvetica,black",
frame=["nswe", "xaf10000", "yaf10000"],
hist=True,
fig=fig,
origin_shift="x",
points=constraint_points[~constraint_points.inside],
points_style="p1p",
)
# add mean / rms to plot
fig.text(
position="BR",
justify="TR",
text=f"RMSE: {round(utils.rmse(dif), 2)}",
fill="white",
font="16p,Helvetica,black",
offset="j0c/.2",
clearance="+tO",
no_clip=True,
)
fig.plot(
x=constraint_points[constraint_points.inside].easting,
y=constraint_points[constraint_points.inside].northing,
style="c.15c",
fill="white",
pen="1p,black",
)
fig.text(
position="TL",
text="b",
fill="white",
pen=True,
font="16p,Helvetica,black",
offset="j.2/.2",
clearance="+tO",
no_clip=True,
)
# plot the true regional misfit
fig = maps.plot_grd(
grav_grid.reg,
fig_height=10,
cpt_lims=reg_lims,
cmap="viridis",
title="Estimated regional misfit",
title_font="16p,Helvetica,black",
cbar_label="mGal",
cbar_font="18p,Helvetica,black",
frame=["nswe", "xaf10000", "yaf10000"],
hist=True,
fig=fig,
origin_shift="x",
points=constraint_points[~constraint_points.inside],
points_style="p1p",
)
fig.plot(
x=constraint_points[constraint_points.inside].easting,
y=constraint_points[constraint_points.inside].northing,
style="c.15c",
fill="white",
pen="1p,black",
)
fig.text(
position="TL",
text="c",
fill="white",
pen=True,
font="16p,Helvetica,black",
offset="j.2/.2",
clearance="+tO",
no_clip=True,
)
fig.show()
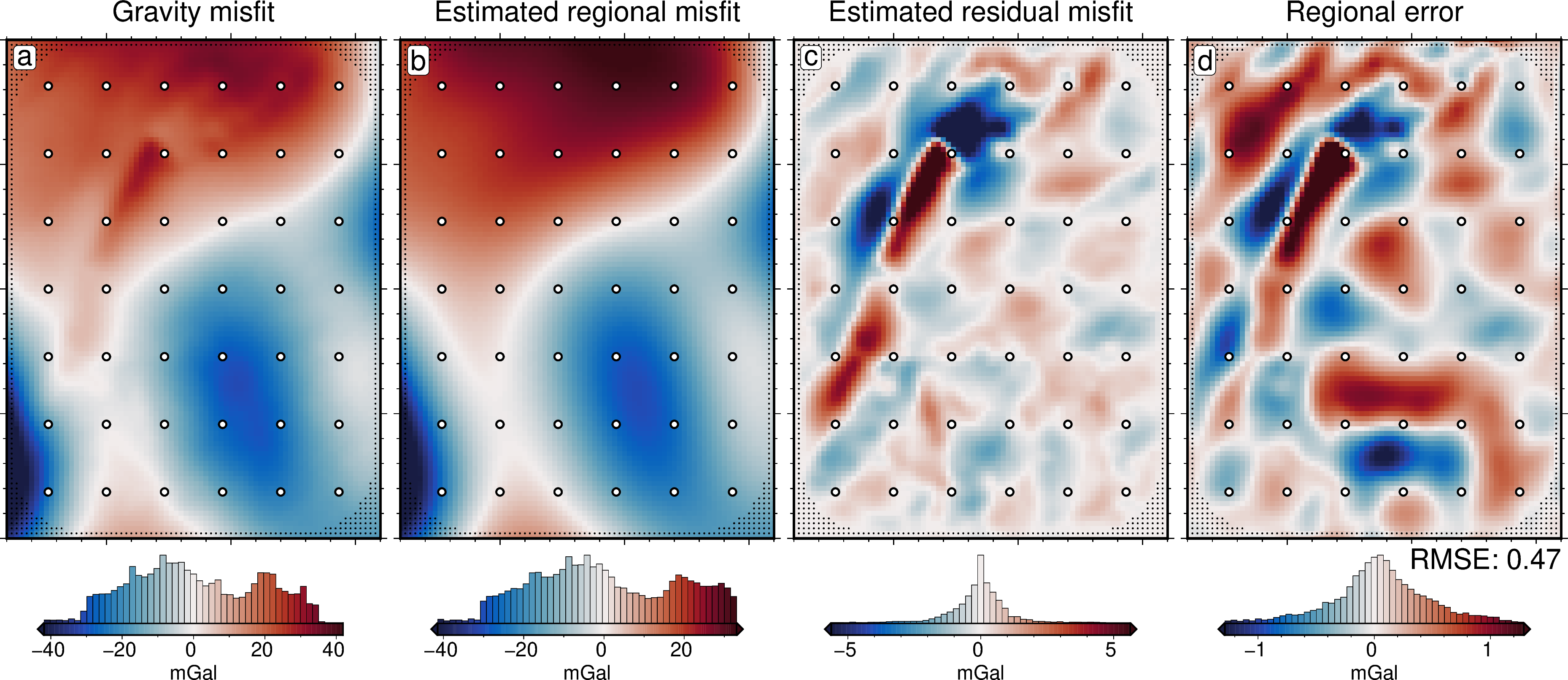
[46]:
# plot gravity misfit
dif = grav_grid.gravity_anomaly - grav_grid.starting_gravity
fig = maps.plot_grd(
dif,
fig_height=10,
cmap="balance+h0",
cpt_lims=polar_utils.get_min_max(
dif,
absolute=True,
robust=True,
robust_percentiles=(0.01, 0.99),
),
title="Gravity misfit",
title_font="16p,Helvetica,black",
cbar_label="mGal",
cbar_font="18p,Helvetica,black",
frame=["nswe", "xaf10000", "yaf10000"],
hist=True,
points=constraint_points[~constraint_points.inside],
points_style="p1p",
)
fig.plot(
x=constraint_points[constraint_points.inside].easting,
y=constraint_points[constraint_points.inside].northing,
style="c.15c",
fill="white",
pen="1p,black",
)
fig.text(
position="TL",
text="a",
fill="white",
pen=True,
font="16p,Helvetica,black",
offset="j.2/.2",
clearance="+tO",
no_clip=True,
)
# plot the estimated regional
reg_lims = polar_utils.get_combined_min_max(
[grav_grid.true_reg, grav_grid.reg],
robust=True,
robust_percentiles=(0.01, 0.99),
)
fig = maps.plot_grd(
grav_grid.reg,
fig_height=10,
cmap="balance+h0",
cpt_lims=reg_lims,
title="Estimated regional misfit",
title_font="16p,Helvetica,black",
cbar_label="mGal",
cbar_font="18p,Helvetica,black",
frame=["nswe", "xaf10000", "yaf10000"],
hist=True,
fig=fig,
origin_shift="x",
points=constraint_points[~constraint_points.inside],
points_style="p1p",
)
fig.plot(
x=constraint_points[constraint_points.inside].easting,
y=constraint_points[constraint_points.inside].northing,
style="c.15c",
fill="white",
pen="1p,black",
)
fig.text(
position="TL",
text="b",
fill="white",
pen=True,
font="16p,Helvetica,black",
offset="j.2/.2",
clearance="+tO",
no_clip=True,
)
# plot the estimated residual
fig = maps.plot_grd(
grav_grid.res,
fig_height=10,
cmap="balance+h0",
cpt_lims=polar_utils.get_min_max(
grav_grid.res,
absolute=True,
robust=True,
robust_percentiles=(0.01, 0.99),
),
title="Estimated residual misfit",
title_font="16p,Helvetica,black",
cbar_label="mGal",
cbar_font="18p,Helvetica,black",
frame=["nswe", "xaf10000", "yaf10000"],
hist=True,
fig=fig,
origin_shift="x",
points=constraint_points[~constraint_points.inside],
points_style="p1p",
)
fig.plot(
x=constraint_points[constraint_points.inside].easting,
y=constraint_points[constraint_points.inside].northing,
style="c.15c",
fill="white",
pen="1p,black",
)
fig.text(
position="TL",
text="c",
fill="white",
pen=True,
font="16p,Helvetica,black",
offset="j.2/.2",
clearance="+tO",
no_clip=True,
)
# plot the regional error
dif = grav_grid.true_reg - grav_grid.reg
fig = maps.plot_grd(
dif,
fig_height=10,
cmap="balance+h0",
cpt_lims=polar_utils.get_min_max(
dif,
absolute=True,
robust=True,
robust_percentiles=(0.01, 0.99),
),
title="Regional error",
title_font="16p,Helvetica,black",
cbar_label="mGal", # f"mGal, RMSE: {round(utils.rmse(dif),2)}",
cbar_font="18p,Helvetica,black",
frame=["nswe", "xaf10000", "yaf10000"],
hist=True,
fig=fig,
origin_shift="x",
points=constraint_points[~constraint_points.inside],
points_style="p1p",
)
# add mean / rms to plot
fig.text(
position="BR",
justify="TR",
text=f"RMSE: {round(utils.rmse(dif), 2)}",
fill="white",
font="16p,Helvetica,black",
offset="j0c/.2",
clearance="+tO",
no_clip=True,
)
fig.plot(
x=constraint_points[constraint_points.inside].easting,
y=constraint_points[constraint_points.inside].northing,
style="c.15c",
fill="white",
pen="1p,black",
)
fig.text(
position="TL",
text="d",
fill="white",
pen=True,
font="16p,Helvetica,black",
offset="j.2/.2",
clearance="+tO",
no_clip=True,
)
fig.show()
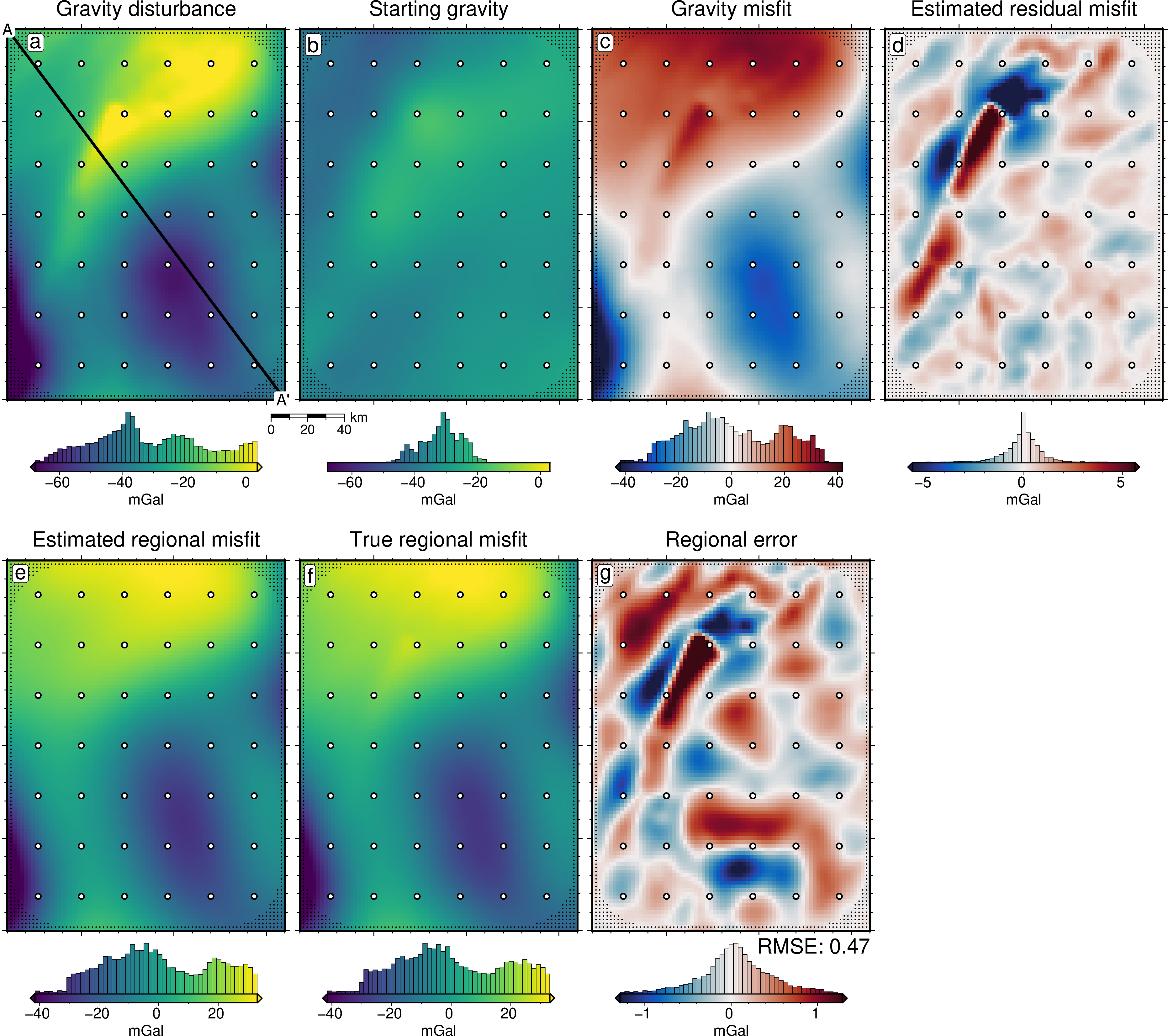
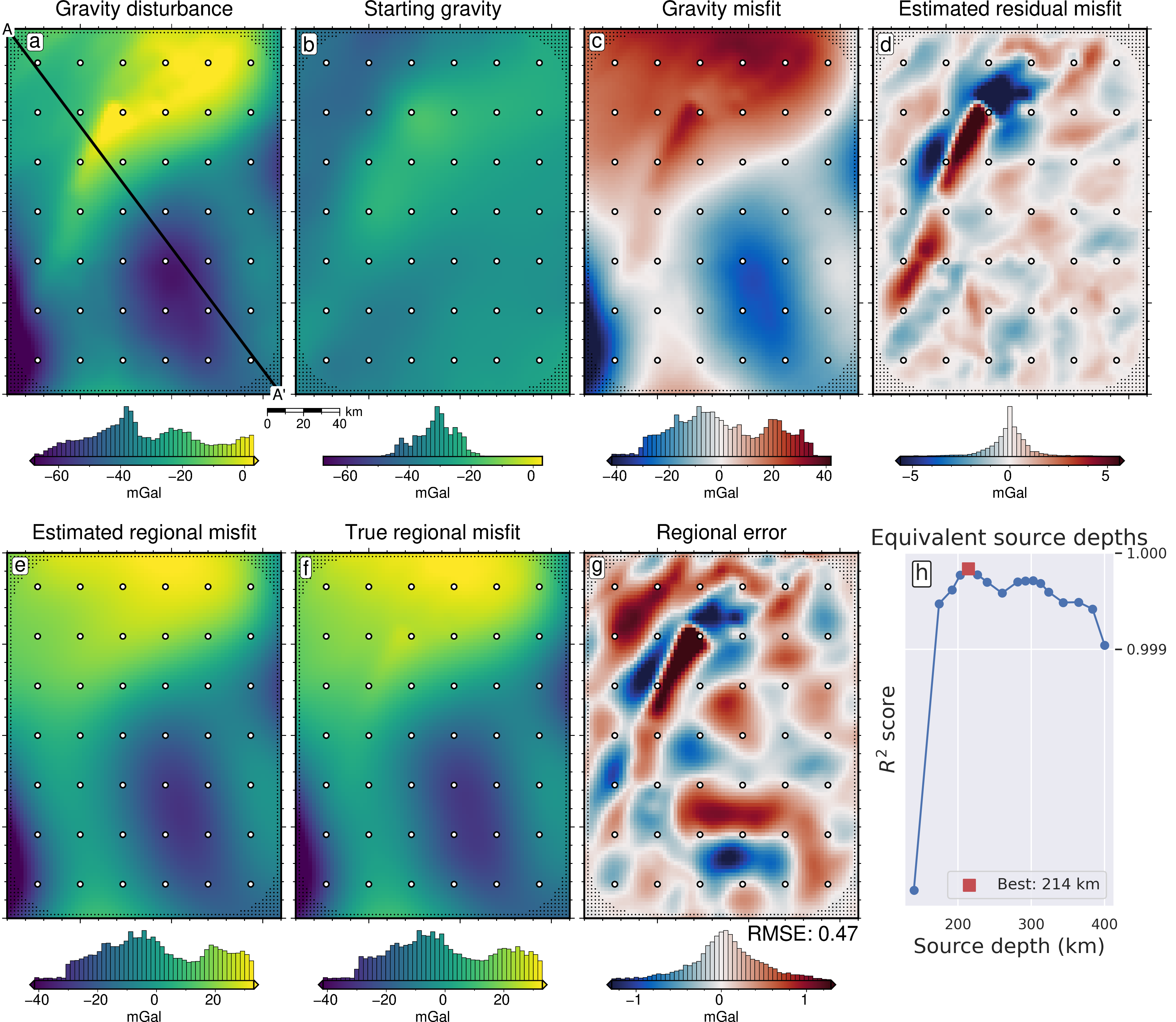
[47]:
lims = polar_utils.get_min_max(
grav_grid.gravity_anomaly,
robust=True,
)
# plot the observed gravity
fig = maps.plot_grd(
grav_grid.gravity_anomaly,
fig_height=10,
cpt_lims=lims,
title="Gravity disturbance",
title_font="16p,Helvetica,black",
cbar_label="mGal",
cbar_font="18p,Helvetica,black",
frame=["nswe", "xaf10000", "yaf10000"],
hist=True,
scalebar=True,
scalebar_position="jBR+o-1.6c/-.6c",
points=constraint_points[~constraint_points.inside],
points_style="p1p",
)
fig.plot(
x=constraint_points[constraint_points.inside].easting,
y=constraint_points[constraint_points.inside].northing,
style="c.15c",
fill="white",
pen="1p,black",
)
# plot profiles location, and endpoints on map
fig.plot(
vd.line_coordinates(profile_start, profile_end, size=100),
pen="2p,black",
)
fig.text(
x=profile_start[0],
y=profile_start[1],
text="A",
fill="white",
font="12p,Helvetica,black",
justify="CM",
clearance="+tO",
no_clip=True,
)
fig.text(
x=profile_end[0],
y=profile_end[1],
text="A' ",
fill="white",
font="12p,Helvetica,black",
justify="CM",
clearance="+tO",
no_clip=True,
)
fig.text(
position="TL",
text="a",
fill="white",
pen=True,
font="16p,Helvetica,black",
offset="j.6/.2",
clearance="+tO",
no_clip=True,
)
# plot the starting gravity
fig = maps.plot_grd(
grav_grid.starting_gravity,
fig_height=10,
cpt_lims=lims,
title="Starting gravity",
title_font="16p,Helvetica,black",
cbar_label="mGal",
cbar_font="18p,Helvetica,black",
frame=["nswe", "xaf10000", "yaf10000"],
hist=True,
fig=fig,
origin_shift="x",
points=constraint_points[~constraint_points.inside],
points_style="p1p",
)
fig.plot(
x=constraint_points[constraint_points.inside].easting,
y=constraint_points[constraint_points.inside].northing,
style="c.15c",
fill="white",
pen="1p,black",
)
fig.text(
position="TL",
text="b",
fill="white",
pen=True,
font="16p,Helvetica,black",
offset="j.2/.2",
clearance="+tO",
no_clip=True,
)
# plot gravity misfit
dif = grav_grid.gravity_anomaly - grav_grid.starting_gravity
fig = maps.plot_grd(
dif,
fig_height=10,
cmap="balance+h0",
cpt_lims=polar_utils.get_min_max(
dif,
absolute=True,
robust=True,
robust_percentiles=(0.01, 0.99),
),
title="Gravity misfit",
title_font="16p,Helvetica,black",
cbar_label="mGal",
cbar_font="18p,Helvetica,black",
frame=["nswe", "xaf10000", "yaf10000"],
hist=True,
fig=fig,
origin_shift="x",
points=constraint_points[~constraint_points.inside],
points_style="p1p",
)
fig.plot(
x=constraint_points[constraint_points.inside].easting,
y=constraint_points[constraint_points.inside].northing,
style="c.15c",
fill="white",
pen="1p,black",
)
fig.text(
position="TL",
text="c",
fill="white",
pen=True,
font="16p,Helvetica,black",
offset="j.2/.2",
clearance="+tO",
no_clip=True,
)
# plot the estimated residual
fig = maps.plot_grd(
grav_grid.res,
fig_height=10,
cmap="balance+h0",
cpt_lims=polar_utils.get_min_max(
grav_grid.res,
absolute=True,
robust=True,
robust_percentiles=(0.01, 0.99),
),
title="Estimated residual misfit",
title_font="16p,Helvetica,black",
cbar_label="mGal",
cbar_font="18p,Helvetica,black",
frame=["nswe", "xaf10000", "yaf10000"],
hist=True,
fig=fig,
origin_shift="x",
points=constraint_points[~constraint_points.inside],
points_style="p1p",
)
fig.plot(
x=constraint_points[constraint_points.inside].easting,
y=constraint_points[constraint_points.inside].northing,
style="c.15c",
fill="white",
pen="1p,black",
)
fig.text(
position="TL",
text="d",
fill="white",
pen=True,
font="16p,Helvetica,black",
offset="j.2/.2",
clearance="+tO",
no_clip=True,
)
# plot the estimated regional
reg_lims = polar_utils.get_combined_min_max(
[grav_grid.true_reg, grav_grid.reg],
robust=True,
robust_percentiles=(0.01, 0.99),
)
fig = maps.plot_grd(
grav_grid.reg,
fig_height=10,
cmap="viridis",
cpt_lims=reg_lims,
title="Estimated regional misfit",
title_font="16p,Helvetica,black",
cbar_label="mGal",
cbar_font="18p,Helvetica,black",
frame=["nswe", "xaf10000", "yaf10000"],
hist=True,
fig=fig,
origin_shift="both",
xshift_amount=-3,
points=constraint_points[~constraint_points.inside],
points_style="p1p",
)
fig.plot(
x=constraint_points[constraint_points.inside].easting,
y=constraint_points[constraint_points.inside].northing,
style="c.15c",
fill="white",
pen="1p,black",
)
fig.text(
position="TL",
text="e",
fill="white",
pen=True,
font="16p,Helvetica,black",
offset="j.2/.2",
clearance="+tO",
no_clip=True,
)
# plot the true regional misfit
fig = maps.plot_grd(
grav_grid.true_reg,
fig_height=10,
cmap="viridis",
cpt_lims=reg_lims,
title="True regional misfit",
title_font="16p,Helvetica,black",
cbar_label="mGal",
cbar_font="18p,Helvetica,black",
frame=["nswe", "xaf10000", "yaf10000"],
hist=True,
fig=fig,
origin_shift="x",
points=constraint_points[~constraint_points.inside],
points_style="p1p",
)
fig.plot(
x=constraint_points[constraint_points.inside].easting,
y=constraint_points[constraint_points.inside].northing,
style="c.15c",
fill="white",
pen="1p,black",
)
fig.text(
position="TL",
text="f",
fill="white",
pen=True,
font="16p,Helvetica,black",
offset="j.2/.2",
clearance="+tO",
no_clip=True,
)
# plot the regional error
dif = grav_grid.true_reg - grav_grid.reg
fig = maps.plot_grd(
dif,
fig_height=10,
cmap="balance+h0",
cpt_lims=polar_utils.get_min_max(
dif,
absolute=True,
robust=True,
robust_percentiles=(0.01, 0.99),
),
title="Regional error",
title_font="16p,Helvetica,black",
cbar_label="mGal", # f"mGal, RMSE: {round(utils.rmse(dif),2)}",
cbar_font="18p,Helvetica,black",
frame=["nswe", "xaf10000", "yaf10000"],
hist=True,
fig=fig,
origin_shift="x",
points=constraint_points[~constraint_points.inside],
points_style="p1p",
)
# add mean / rms to plot
fig.text(
position="BR",
justify="TR",
text=f"RMSE: {round(utils.rmse(dif), 2)}",
fill="white",
font="16p,Helvetica,black",
offset="j0c/.2",
clearance="+tO",
no_clip=True,
)
fig.plot(
x=constraint_points[constraint_points.inside].easting,
y=constraint_points[constraint_points.inside].northing,
style="c.15c",
fill="white",
pen="1p,black",
)
fig.text(
position="TL",
text="g",
fill="white",
pen=True,
font="16p,Helvetica,black",
offset="j.2/.2",
clearance="+tO",
no_clip=True,
)
fig.show()
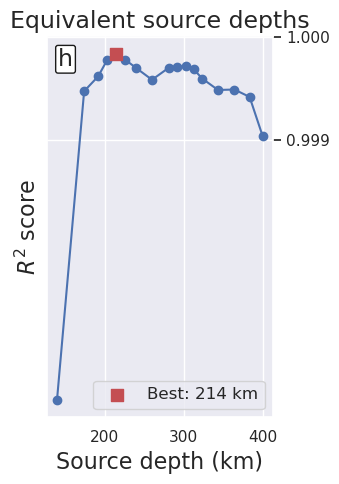
[48]:
df = regional_study.trials_dataframe().sort_values("params_depth")
df = df.iloc[3:].reset_index(drop=True)
_fig, ax = plt.subplots(figsize=(3.5, 5))
ax.plot(
df.params_depth / 1e3,
df.value,
marker="o",
)
best_ind = df.value.idxmax()
ax.scatter(
x=df.iloc[best_ind].params_depth / 1e3,
y=df.iloc[best_ind].value,
marker="s",
color="r",
s=80,
zorder=20,
label=f"Best: {round(df.iloc[best_ind].params_depth / 1e3)} km",
)
ax.legend(fontsize=12)
ax.set_xlabel("Source depth (km)", fontsize=16)
ax.set_ylabel("$R^2$ score", fontsize=16)
ax.set_title("Equivalent source depths", fontsize=17)
ax.annotate(
"h",
xy=(0.05, 0.97),
xycoords="axes fraction",
fontsize=17,
va="top",
ha="left",
bbox=dict(boxstyle="round,pad=0.1", facecolor="white", edgecolor="k", pad=3.0),
)
ax.set_yticks([0.999, 1])
ax.yaxis.tick_right()
ax.yaxis.set_major_formatter(mpl.ticker.FormatStrFormatter("%.3f"))
_fig.tight_layout()
_fig.savefig(
f"../paper/figures/Ross_Sea_0{i}_eq_sources.png", dpi=300, bbox_inches="tight"
)
[49]:
fig.image(
imagefile=f"../paper/figures/Ross_Sea_0{i}_eq_sources.png",
position="n1.01/.48+jML",
)
fig.savefig(f"../paper/figures/Ross_Sea_0{i}_grav_anomalies.png")
fig.show()
[20]:
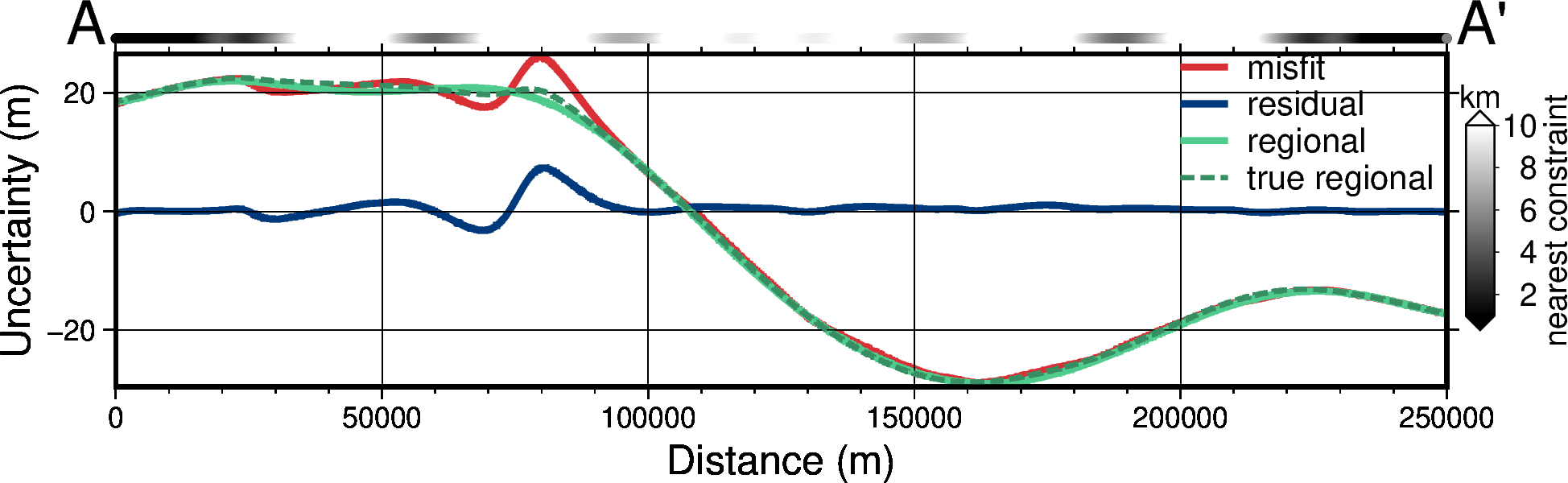
titles = [
"misfit",
"residual",
"regional",
"true regional",
]
grids = [
grav_grid.misfit,
grav_grid.res,
grav_grid.reg,
grav_grid.true_reg,
]
data_ylims = None
data_dict = profiles.make_data_dict(
names=titles,
grids=grids,
colors=[ # https://www.simplifiedsciencepublishing.com/resources/best-color-palettes-for-scientific-figures-and-data-visualizations
"216/48/52", # red, d83034
"0/58/125", # dark blue, 003a7d
"78/203/141", # green, 4ecb8d
f"{78 * 0.7}/{203 * 0.7}/{141 * 0.7}", # green, darker than above
# "199/1/255", # purple, c701ff
# "255/157/58", # orange, ff9d3a
# "0/141/255", # med blue, 008dff
# "255/115/182", # pink, ff73b6
# "249/232/88", # yellow, f9e858
],
)
fig, df_data = profiles.plot_data(
"points",
start=profile_start,
stop=profile_end,
num=10000,
fig_height=4,
fig_width=14,
data_dict=data_dict,
data_legend_loc="jTR+o0c/-.2c",
data_buffer=0.01,
data_frame=["neSW", "xafg+lDistance (m)", "yag+lUncertainty (m)"],
data_pen_style=[None, None, None, "4_2:2p"],
data_pen_thickness=[2, 2, 2, 1.5],
share_yaxis=True,
start_label="A",
end_label="A' ",
)
# get grid of min dists to nearest constraints
min_dist = utils.normalized_mindist(
constraint_points,
grids[0],
)
# set y height in data units for add dist to constraint points
data_max = df_data.drop(columns=["easting", "northing", "dist"]).max().max()
if data_ylims is not None:
data_max = data_ylims[1]
df_data = profiles.sample_grids(
df_data, min_dist, interpolation="b", sampled_name="min_dist"
)
# set color limits
series = [1e3, 10e3]
pygmt.makecpt(
cmap="gray",
series=series,
background=True,
)
fig.shift_origin(
yshift=".2c",
)
fig.plot(
x=df_data.dist,
y=np.ones_like(df_data.dist) * data_max,
style="c0.1c",
fill=df_data.min_dist,
cmap=True,
no_clip=True,
)
fig.shift_origin(
yshift="-.2c",
)
with pygmt.config(
FONT="26p,Helvetica,black",
):
fig.colorbar(
cmap=True,
position="jMR+w2c/.3c+jML+v+o0.2c/0c+e",
frame=["xa+lnearest constraint", "y+lkm"],
scale=0.001,
)
fig.savefig(f"../paper/figures/Ross_Sea_0{i}_grav_anomalies_profiles.png")
fig.show()
grdtrack [WARNING]: Some input points were outside the grid domain(s).
grdtrack [WARNING]: Some input points were outside the grid domain(s).
grdtrack [WARNING]: Some input points were outside the grid domain(s).
grdtrack [WARNING]: Some input points were outside the grid domain(s).
grdtrack [WARNING]: Some input points were outside the grid domain(s).
[51]:
np.abs(dif).mean(), np.abs(dif).max()
[51]:
(<xarray.DataArray ()> Size: 8B
array(0.33686614),
<xarray.DataArray ()> Size: 8B
array(2.7549579))
[ ]:
titles = [
"True error",
"Total uncertainty",
"Uncert. from damping",
"Uncert. from density",
]
grids = [
np.abs(stochastic_mean - bathymetry),
sensitivity_ds.full_stdev,
sensitivity_ds.damping_stdev,
sensitivity_ds.density_stdev,
]
sensitivity_plot(
titles,
grids,
constraint_points=constraint_points,
num_in_top_row=4,
# fname=f"../paper/figures/Ross_Sea_0{i}_sensitivity.png",
)
gmtset [WARNING]: Representation of font type not recognized. Using default.
gmtset [WARNING]: Representation of font type not recognized. Using default.
gmtset [WARNING]: Representation of font type not recognized. Using default.
gmtset [WARNING]: Representation of font type not recognized. Using default.
[53]:
err = final_bathymetry - bathymetry
fig = maps.plot_grd(
err,
fig_height=10,
cmap="balance+h0",
cpt_lims=polar_utils.get_min_max(
err,
absolute=True,
robust=True,
robust_percentiles=(0.01, 0.99),
),
title="True error",
title_font="16p,Helvetica,black",
cbar_label="mGal",
cbar_font="18p,Helvetica,black",
frame=["nswe", "xaf10000", "yaf10000"],
hist=True,
)
fig.show()
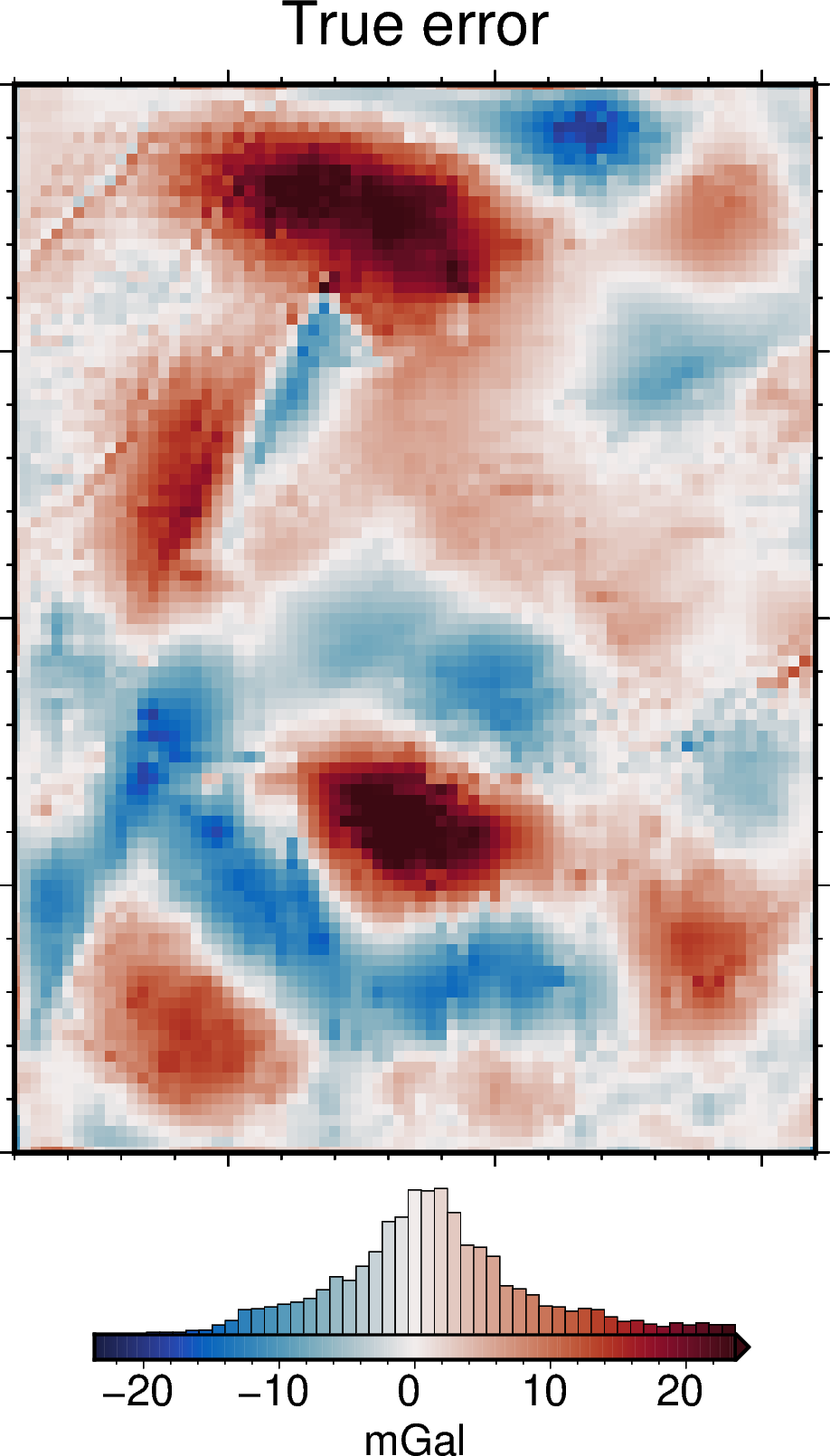
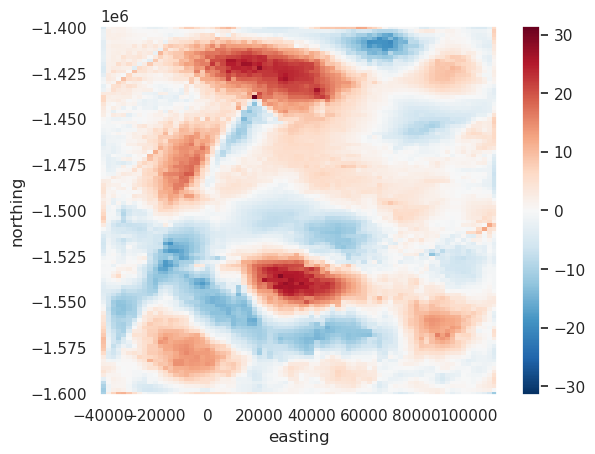
[54]:
err.plot()
[54]:
<matplotlib.collections.QuadMesh at 0x7fa7f45c1340>
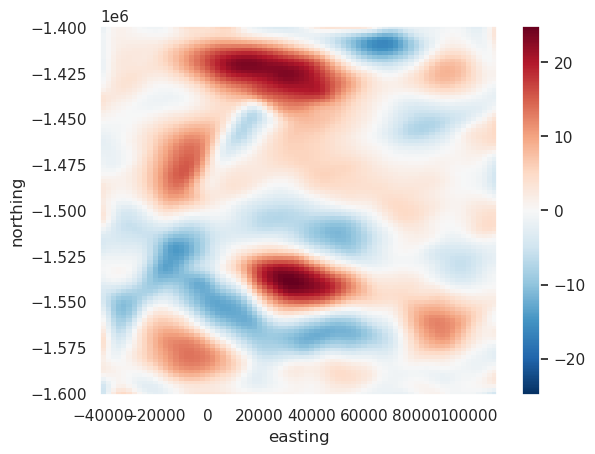
[55]:
filt = utils.filter_grid(err, 20e3)
filt.plot()
[55]:
<matplotlib.collections.QuadMesh at 0x7fa7f4192f90>
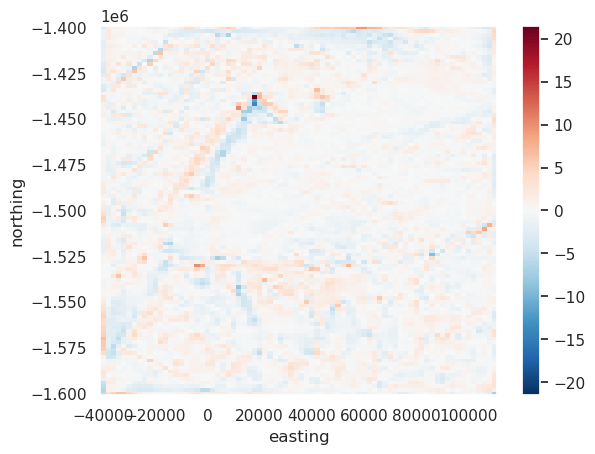
[56]:
filt2 = utils.filter_grid(err, 20e3, filt_type="highpass")
filt2.plot()
[56]:
<matplotlib.collections.QuadMesh at 0x7fa7f4594620>
5: Realistic scenario¶
[23]:
i = 5
final_bathymetry = final_bathymetries[i - 1]
inversion_results = optimal_results[i - 1]
damping_study = damping_studies[i - 1]
density_study = density_studies[i - 1]
with pathlib.Path(
f"../tmp_outputs/Ross_Sea_0{i}_regional_sep_true_density.pickle"
).open("rb") as f:
regional_study = pickle.load(f)
constraint_points = pd.read_csv(
f"../results/Ross_Sea/Ross_Sea_0{i}_constraint_points.csv"
)
uncertainty = final_uncertainties[i - 1]
stochastic_mean = final_stochastic_means[i - 1]
grav_df = pd.read_csv(f"../results/Ross_Sea/Ross_Sea_0{i}_grav_df_true_density.csv")
eqs_uncert = pd.read_csv(f"../results/Ross_Sea/Ross_Sea_0{i}_grav_df.csv")["eqs_uncert"]
grav_df["eqs_uncert"] = eqs_uncert
sensitivity_ds = sensitivity_results[i - 1]
[24]:
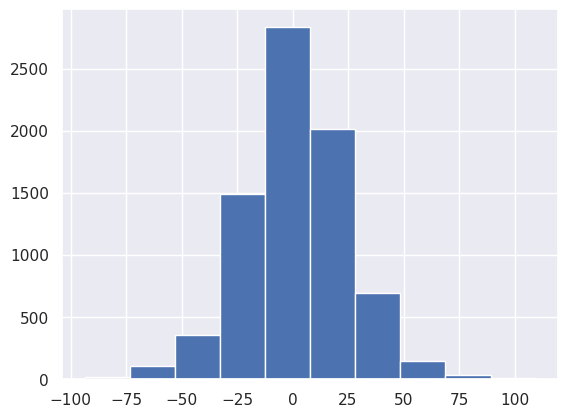
(final_bathymetry - bathymetry).plot.hist()
[24]:
(array([ 13., 105., 352., 1487., 2836., 2011., 690., 146., 32.,
4.]),
array([-93.60144085, -73.29190458, -52.98236831, -32.67283205,
-12.36329578, 7.94624049, 28.25577676, 48.56531303,
68.87484929, 89.18438556, 109.49392183]),
<BarContainer object of 10 artists>)
[59]:
(starting_bathymetry - bathymetry).to_numpy().max()
[59]:
np.float64(164.39923609925518)
[60]:
(final_bathymetry - bathymetry).to_numpy().max()
[60]:
np.float64(109.49392183017909)
[61]:
grav_df.head()
[61]:
| northing | easting | upward | bathymetry_grav | basement_grav_normalized | basement_grav | gravity_anomaly_full_res_no_noise | gravity_anomaly_full_res | uncert | eqs_gravity_anomaly | gravity_anomaly | starting_gravity | true_res | misfit | reg | res | reg_uncert | eqs_uncert | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | -1600000.0 | -40000.0 | 1000.0 | -35.551085 | -0.575645 | -36.300394 | -71.851479 | -72.911198 | 3 | -64.481389 | -64.481389 | -35.578561 | -3.009718 | -28.902828 | -29.704963 | 0.802136 | 0.314038 | 1.537731 |
| 1 | -1600000.0 | -38000.0 | 1000.0 | -36.054683 | -0.523223 | -33.941405 | -69.996088 | -72.483909 | 3 | -64.163085 | -64.163085 | -36.042642 | -3.088852 | -28.120443 | -28.604746 | 0.484303 | 0.228852 | 1.318696 |
| 2 | -1600000.0 | -36000.0 | 1000.0 | -36.473168 | -0.466365 | -31.382798 | -67.855967 | -68.870245 | 3 | -63.178870 | -63.178870 | -36.460095 | -3.125521 | -26.718775 | -27.043474 | 0.324699 | 0.287741 | 1.168135 |
| 3 | -1600000.0 | -34000.0 | 1000.0 | -36.755627 | -0.406022 | -28.667352 | -65.422980 | -61.536475 | 3 | -61.636021 | -61.636021 | -36.784649 | -3.111132 | -24.851373 | -25.130963 | 0.279591 | 0.298262 | 0.998435 |
| 4 | -1600000.0 | -32000.0 | 1000.0 | -36.951045 | -0.343163 | -25.838709 | -62.789754 | -65.528788 | 3 | -59.623519 | -59.623519 | -36.998469 | -3.110982 | -22.625050 | -22.965266 | 0.340216 | 0.269691 | 0.823200 |
[25]:
grav_error = np.abs(grav_df.gravity_anomaly_full_res_no_noise - grav_df.gravity_anomaly)
print(
f"MAE of gravity: {np.abs(grav_df.gravity_anomaly_full_res_no_noise - grav_df.gravity_anomaly).mean()}"
)
print(
f"RMSE of gravity: {utils.rmse(grav_df.gravity_anomaly_full_res_no_noise - grav_df.gravity_anomaly)}"
)
grav_error.describe()
MAE of gravity: 0.8472477774320722
RMSE of gravity: 1.135396522721251
[25]:
count 7676.000000
mean 0.847248
std 0.755891
min 0.000369
25% 0.302118
50% 0.651053
75% 1.179984
max 8.188145
dtype: float64
[26]:
# calculate the true residual and regional misfit
grav_df["true_res"] = grav_df.bathymetry_grav - grav_df.starting_gravity
grav_df["true_reg"] = grav_df.basement_grav
grav_df.head()
[26]:
| northing | easting | upward | bathymetry_grav | basement_grav_normalized | basement_grav | gravity_anomaly_full_res_no_noise | gravity_anomaly_full_res | uncert | eqs_gravity_anomaly | gravity_anomaly | starting_gravity | true_res | misfit | reg | res | reg_uncert | eqs_uncert | true_reg | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | -1600000.0 | -40000.0 | 1000.0 | -35.551085 | -0.575645 | -36.300394 | -71.851479 | -72.911198 | 3 | -64.481389 | -64.481389 | -35.578561 | 0.027476 | -28.902828 | -29.704963 | 0.802136 | 0.314038 | 1.537731 | -36.300394 |
| 1 | -1600000.0 | -38000.0 | 1000.0 | -36.054683 | -0.523223 | -33.941405 | -69.996088 | -72.483909 | 3 | -64.163085 | -64.163085 | -36.042642 | -0.012041 | -28.120443 | -28.604746 | 0.484303 | 0.228852 | 1.318696 | -33.941405 |
| 2 | -1600000.0 | -36000.0 | 1000.0 | -36.473168 | -0.466365 | -31.382798 | -67.855967 | -68.870245 | 3 | -63.178870 | -63.178870 | -36.460095 | -0.013073 | -26.718775 | -27.043474 | 0.324699 | 0.287741 | 1.168135 | -31.382798 |
| 3 | -1600000.0 | -34000.0 | 1000.0 | -36.755627 | -0.406022 | -28.667352 | -65.422980 | -61.536475 | 3 | -61.636021 | -61.636021 | -36.784649 | 0.029021 | -24.851373 | -25.130963 | 0.279591 | 0.298262 | 0.998435 | -28.667352 |
| 4 | -1600000.0 | -32000.0 | 1000.0 | -36.951045 | -0.343163 | -25.838709 | -62.789754 | -65.528788 | 3 | -59.623519 | -59.623519 | -36.998469 | 0.047423 | -22.625050 | -22.965266 | 0.340216 | 0.269691 | 0.823200 | -25.838709 |
[27]:
print(f"MA of regional: {np.abs(grav_df.reg).mean()}")
print(f"RMS of regional: {utils.rmse(grav_df.reg)}")
print(f"MAE of regional: {np.abs(grav_df.true_reg - grav_df.reg).mean()}")
print(f"RMSE of regional: {utils.rmse(grav_df.true_reg - grav_df.reg)}")
print(f"Max regional error: {np.abs(grav_df.true_reg - grav_df.reg).max()}")
MA of regional: 15.163261653873885
RMS of regional: 18.085531433319662
MAE of regional: 0.8972725248045438
RMSE of regional: 1.1826080946741815
Max regional error: 7.489381481647513
[28]:
grav_grid = grav_df.set_index(["northing", "easting"]).to_xarray()
[66]:
lims = polar_utils.get_min_max(grav_grid.gravity_anomaly_full_res_no_noise)
# plot the noise-free gravity
fig = maps.plot_grd(
grav_grid.gravity_anomaly_full_res_no_noise,
fig_height=10,
cpt_lims=lims,
title="True gravity",
title_font="18p,Helvetica,black",
cbar_label="mGal",
cbar_font="18p,Helvetica,black",
frame=["nswe", "xaf10000", "yaf10000"],
hist=True,
scalebar=True,
scalebar_position="jBR+o-.9c/-.6c",
)
# plot observation points
fig.plot(
grav_survey_df[["easting", "northing"]],
style="c.02c",
fill="black",
)
fig.text(
position="TL",
text="a",
fill="white",
pen=True,
font="16p,Helvetica,black",
offset="j.2/.2",
clearance="+tO",
no_clip=True,
)
# plot the difference between noisy and filtered gravity
dif = grav_grid.gravity_anomaly_full_res_no_noise - grav_grid.gravity_anomaly
fig = maps.plot_grd(
dif,
fig_height=10,
cmap="balance+h0",
cpt_lims=polar_utils.get_min_max(
dif,
absolute=True,
robust=True,
robust_percentiles=(0.01, 0.99),
),
title="Data loss",
title_font="18p,Helvetica,black",
cbar_label="mGal",
cbar_font="18p,Helvetica,black",
frame=["nswe", "xaf10000", "yaf10000"],
hist=True,
fig=fig,
origin_shift="x",
)
# add mean / rms to plot
fig.text(
position="BR",
justify="TR",
text=f"RMS: {round(utils.rmse(dif), 2)}",
fill="white",
font="16p,Helvetica,black",
offset="j0c/.2",
clearance="+tO",
no_clip=True,
)
fig.text(
position="TL",
text="b",
fill="white",
pen=True,
font="16p,Helvetica,black",
offset="j.2/.2",
clearance="+tO",
no_clip=True,
)
# plot the sampled, interpolated, and filtered gravity
fig = maps.plot_grd(
grav_grid.gravity_anomaly,
fig_height=10,
cpt_lims=lims,
title="Observed gravity",
title_font="18p,Helvetica,black",
cbar_label="mGal",
cbar_font="18p,Helvetica,black",
frame=["nswe", "xaf10000", "yaf10000"],
hist=True,
fig=fig,
origin_shift="xshift",
)
fig.text(
position="TL",
text="c",
fill="white",
pen=True,
font="16p,Helvetica,black",
offset="j.2/.2",
clearance="+tO",
no_clip=True,
)
# plot the sampled, interpolated, and filtered gravity
fig = maps.plot_grd(
grav_grid.eqs_uncert,
fig_height=10,
cpt_lims=(
0,
polar_utils.get_min_max(
grav_grid.eqs_uncert,
robust=True,
)[1],
),
cmap="ice",
reverse_cpt=True,
title="Uncertainty", #: RMS: {round(utils.rmse(grav_grid.eqs_uncert),2)}",
title_font="18p,Helvetica,black",
cbar_label="mGal",
cbar_font="18p,Helvetica,black",
frame=["nswe", "xaf10000", "yaf10000"],
hist=True,
fig=fig,
origin_shift="x",
)
# add mean / rms to plot
fig.text(
position="BR",
justify="TR",
text=f"RMS: {round(utils.rmse(grav_grid.eqs_uncert), 2)}",
fill="white",
font="16p,Helvetica,black",
offset="j0c/.2",
clearance="+tO",
no_clip=True,
)
fig.text(
position="TL",
text="d",
fill="white",
pen=True,
font="16p,Helvetica,black",
offset="j.2/.2",
clearance="+tO",
no_clip=True,
)
fig.show()
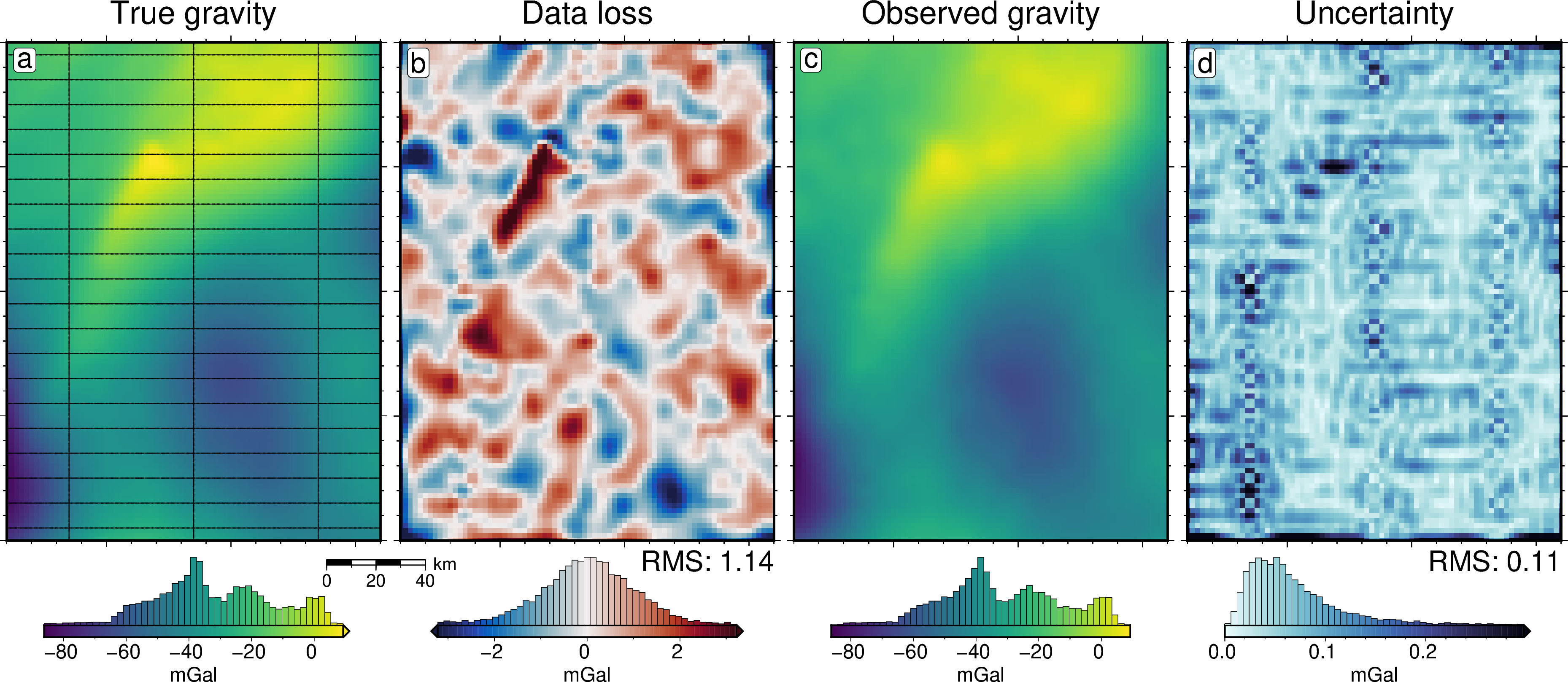
[67]:
filter_widths_df = pd.read_csv(
f"../results/Ross_Sea/Ross_Sea_0{i}_filter_width_rmse.csv"
)
[68]:
_fig, ax = plt.subplots(figsize=(6, 2))
ax.plot(
filter_widths_df.filt_width / 1e3,
filter_widths_df.rmse,
marker="o",
)
best_ind = filter_widths_df.rmse.idxmin()
ax.scatter(
x=filter_widths_df.iloc[best_ind].filt_width / 1e3,
y=filter_widths_df.iloc[best_ind].rmse,
marker="s",
color="r",
s=80,
zorder=20,
label=f"Best: {round(filter_widths_df.iloc[best_ind].filt_width / 1e3, 2)} km",
)
ax.legend(fontsize=14)
ax.set_xlabel("Filter width (km)", fontsize=16)
ax.set_ylabel("RMSE (mGal)", fontsize=16)
ax.set_title("e) Optimal filter width", fontsize=18)
_fig.savefig(
f"../paper/figures/Ross_Sea_0{i}_filters.png", dpi=300, bbox_inches="tight"
)
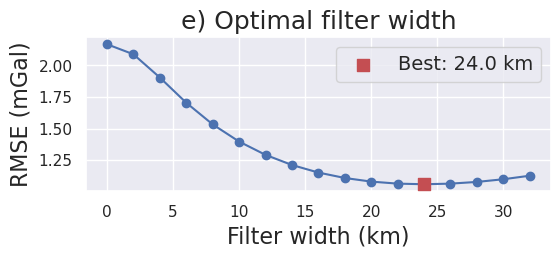
[69]:
fig.image(
imagefile=f"../paper/figures/Ross_Sea_0{i}_filters.png",
position="n-2.1/-.3+jTL",
)
fig.savefig(f"../paper/figures/Ross_Sea_0{i}_grav_error.png")
fig.show()
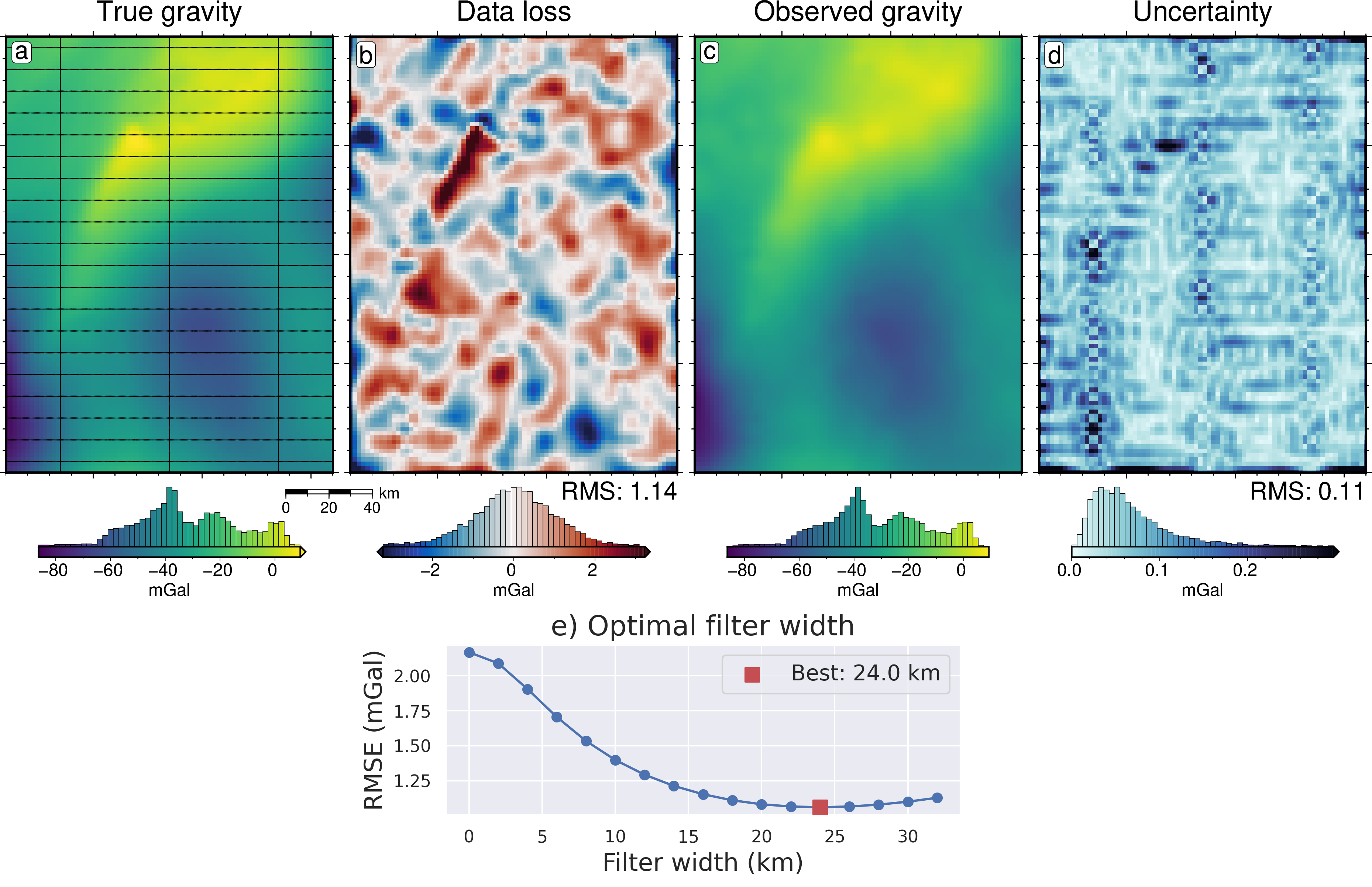
[70]:
reg_lims = polar_utils.get_combined_min_max(
[grav_grid.true_reg, grav_grid.reg],
robust=True,
robust_percentiles=(0.01, 0.99),
)
fig = maps.plot_grd(
grav_grid.true_reg,
fig_height=10,
cmap="viridis",
cpt_lims=reg_lims,
title="True regional misfit",
title_font="16p,Helvetica,black",
cbar_label="mGal",
cbar_font="18p,Helvetica,black",
frame=["nswe", "xaf10000", "yaf10000"],
hist=True,
points=constraint_points[~constraint_points.inside],
points_style="p1p",
scalebar=True,
scalebar_position="jBR+o-.9c/-.6c",
)
fig.plot(
x=constraint_points[constraint_points.inside].easting,
y=constraint_points[constraint_points.inside].northing,
style="c.15c",
fill="white",
pen="1p,black",
)
fig.text(
position="TL",
text="a",
fill="white",
pen=True,
font="16p,Helvetica,black",
offset="j.6/.2",
clearance="+tO",
no_clip=True,
)
# plot the regional error
dif = grav_grid.true_reg - grav_grid.reg
fig = maps.plot_grd(
dif,
fig_height=10,
cmap="balance+h0",
cpt_lims=polar_utils.get_min_max(
dif,
absolute=True,
robust=True,
robust_percentiles=(0.01, 0.99),
),
title="Error",
title_font="16p,Helvetica,black",
cbar_label="mGal", # f"mGal, RMSE: {round(utils.rmse(dif),2)}",
cbar_font="18p,Helvetica,black",
frame=["nswe", "xaf10000", "yaf10000"],
hist=True,
fig=fig,
origin_shift="x",
points=constraint_points[~constraint_points.inside],
points_style="p1p",
)
# add mean / rms to plot
fig.text(
position="BR",
justify="TR",
text=f"RMSE: {round(utils.rmse(dif), 2)}",
fill="white",
font="16p,Helvetica,black",
offset="j0c/.2",
clearance="+tO",
no_clip=True,
)
fig.plot(
x=constraint_points[constraint_points.inside].easting,
y=constraint_points[constraint_points.inside].northing,
style="c.15c",
fill="white",
pen="1p,black",
)
fig.text(
position="TL",
text="b",
fill="white",
pen=True,
font="16p,Helvetica,black",
offset="j.2/.2",
clearance="+tO",
no_clip=True,
)
# plot the true regional misfit
fig = maps.plot_grd(
grav_grid.reg,
fig_height=10,
cpt_lims=reg_lims,
cmap="viridis",
title="Estimated regional misfit",
title_font="16p,Helvetica,black",
cbar_label="mGal",
cbar_font="18p,Helvetica,black",
frame=["nswe", "xaf10000", "yaf10000"],
hist=True,
fig=fig,
origin_shift="x",
points=constraint_points[~constraint_points.inside],
points_style="p1p",
)
fig.plot(
x=constraint_points[constraint_points.inside].easting,
y=constraint_points[constraint_points.inside].northing,
style="c.15c",
fill="white",
pen="1p,black",
)
fig.text(
position="TL",
text="c",
fill="white",
pen=True,
font="16p,Helvetica,black",
offset="j.2/.2",
clearance="+tO",
no_clip=True,
)
fig.show()
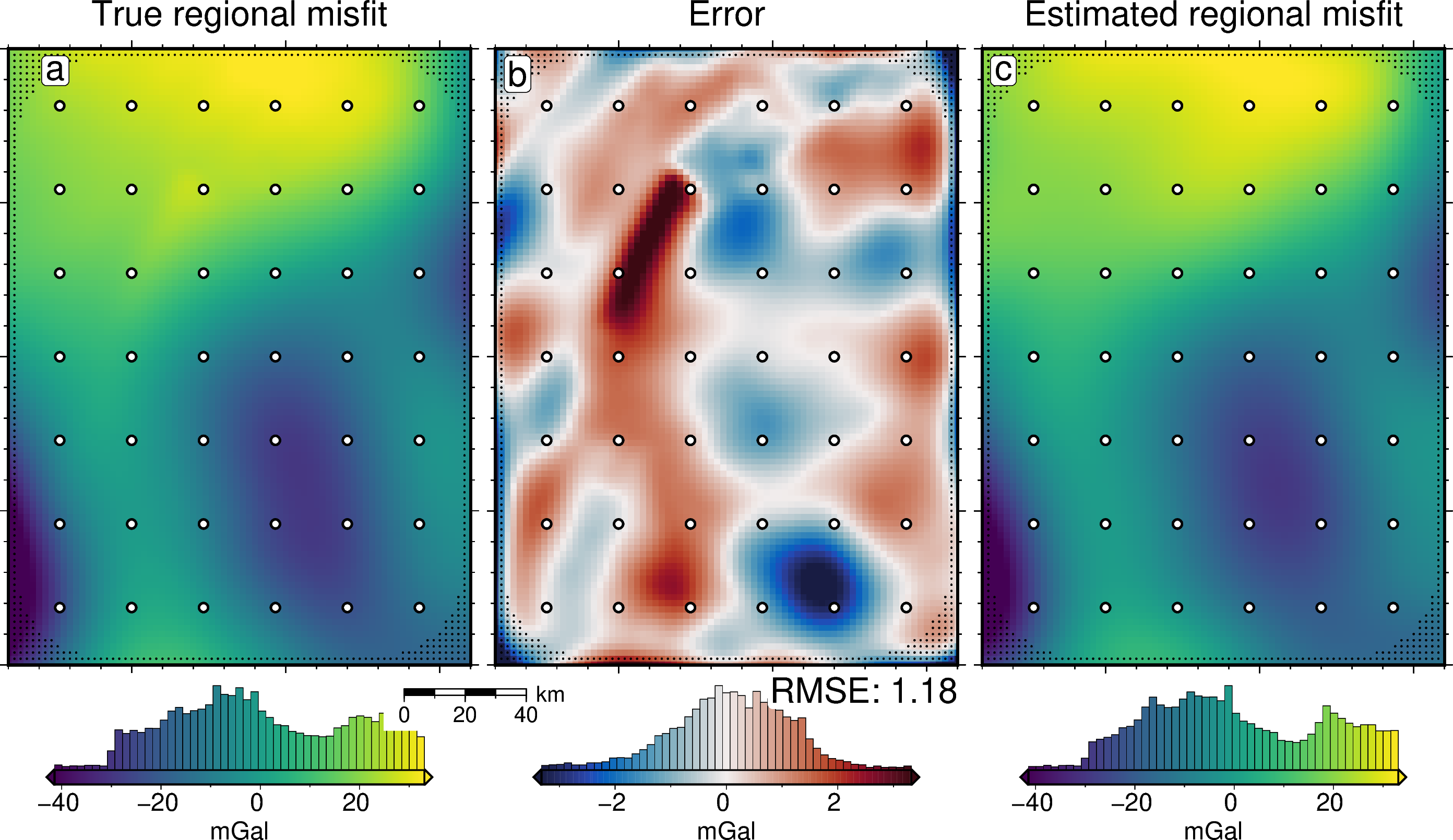
[71]:
reg_lims = polar_utils.get_combined_min_max(
[grav_grid.true_reg, grav_grid.reg],
robust=True,
robust_percentiles=(0.01, 0.99),
)
fig = maps.plot_grd(
grav_grid.true_reg,
fig_height=10,
cmap="viridis",
cpt_lims=reg_lims,
title="True regional misfit",
title_font="16p,Helvetica,black",
cbar_label="mGal",
cbar_font="18p,Helvetica,black",
frame=["nswe", "xaf10000", "yaf10000"],
hist=True,
points=constraint_points[~constraint_points.inside],
points_style="p1p",
scalebar=True,
scalebar_position="jBR+o-.9c/-.6c",
)
fig.plot(
x=constraint_points[constraint_points.inside].easting,
y=constraint_points[constraint_points.inside].northing,
style="c.15c",
fill="white",
pen="1p,black",
)
# plot profiles location, and endpoints on map
fig.plot(
vd.line_coordinates(profile_start, profile_end, size=100),
pen="2p,black",
)
fig.text(
x=profile_start[0],
y=profile_start[1],
text="A",
fill="white",
font="12p,Helvetica,black",
justify="CM",
clearance="+tO",
no_clip=True,
)
fig.text(
x=profile_end[0],
y=profile_end[1],
text="A' ",
fill="white",
font="12p,Helvetica,black",
justify="CM",
clearance="+tO",
no_clip=True,
)
fig.text(
position="TL",
text="a",
fill="white",
pen=True,
font="16p,Helvetica,black",
offset="j.6/.2",
clearance="+tO",
no_clip=True,
)
# plot the regional error
dif = grav_grid.true_reg - grav_grid.reg
fig = maps.plot_grd(
dif,
fig_height=10,
cmap="balance+h0",
cpt_lims=polar_utils.get_min_max(
dif,
absolute=True,
robust=True,
robust_percentiles=(0.01, 0.99),
),
title="Error",
title_font="16p,Helvetica,black",
cbar_label="mGal", # f"mGal, RMSE: {round(utils.rmse(dif),2)}",
cbar_font="18p,Helvetica,black",
frame=["nswe", "xaf10000", "yaf10000"],
hist=True,
fig=fig,
origin_shift="x",
points=constraint_points[~constraint_points.inside],
points_style="p1p",
)
# add mean / rms to plot
fig.text(
position="BR",
justify="TR",
text=f"RMSE: {round(utils.rmse(dif), 2)}",
fill="white",
font="16p,Helvetica,black",
offset="j0c/.2",
clearance="+tO",
no_clip=True,
)
fig.plot(
x=constraint_points[constraint_points.inside].easting,
y=constraint_points[constraint_points.inside].northing,
style="c.15c",
fill="white",
pen="1p,black",
)
fig.text(
position="TL",
text="b",
fill="white",
pen=True,
font="16p,Helvetica,black",
offset="j.2/.2",
clearance="+tO",
no_clip=True,
)
# plot the true regional misfit
fig = maps.plot_grd(
grav_grid.reg,
fig_height=10,
cpt_lims=reg_lims,
cmap="viridis",
title="Estimated regional misfit",
title_font="16p,Helvetica,black",
cbar_label="mGal",
cbar_font="18p,Helvetica,black",
frame=["nswe", "xaf10000", "yaf10000"],
hist=True,
fig=fig,
origin_shift="x",
points=constraint_points[~constraint_points.inside],
points_style="p1p",
)
fig.plot(
x=constraint_points[constraint_points.inside].easting,
y=constraint_points[constraint_points.inside].northing,
style="c.15c",
fill="white",
pen="1p,black",
)
fig.text(
position="TL",
text="c",
fill="white",
pen=True,
font="16p,Helvetica,black",
offset="j.2/.2",
clearance="+tO",
no_clip=True,
)
fig.show()
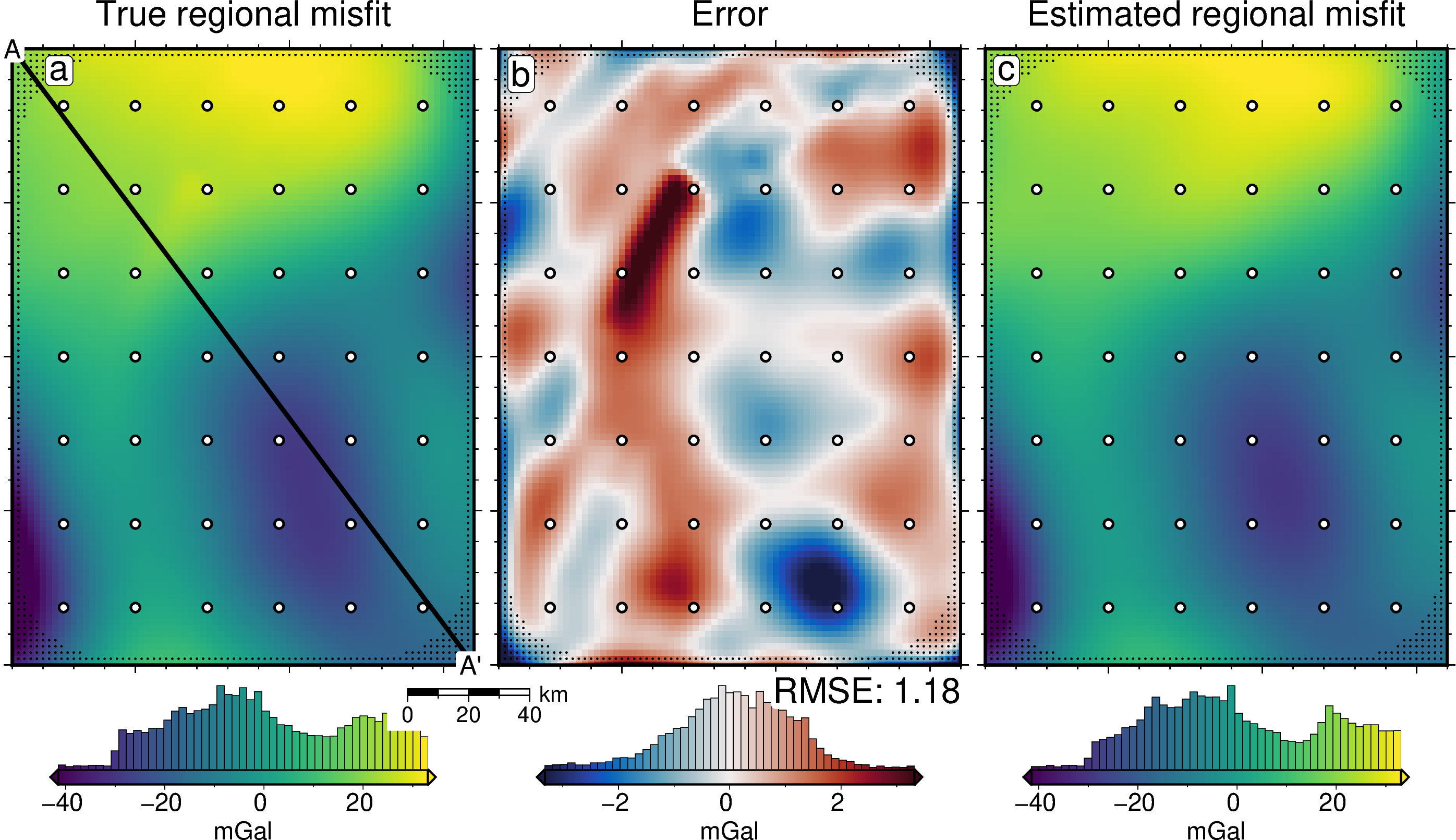
[72]:
df = regional_study.trials_dataframe().sort_values("params_depth")
print(df)
df = df.iloc[2:].reset_index(drop=True)
_fig, ax = plt.subplots(figsize=(3.5, 5))
ax.plot(
df.params_depth / 1e3,
df.value,
marker="o",
)
best_ind = df.value.idxmax()
ax.scatter(
x=df.iloc[best_ind].params_depth / 1e3,
y=df.iloc[best_ind].value,
marker="s",
color="r",
s=80,
zorder=20,
label=f"Best: {round(df.iloc[best_ind].params_depth / 1e3)} km",
)
ax.legend(fontsize=12)
ax.set_xlabel("Source depth (km)", fontsize=16)
ax.set_ylabel("$R^2$ score", fontsize=16)
ax.set_title("Equivalent source depths", fontsize=17)
ax.annotate(
"d",
xy=(0.05, 0.97),
xycoords="axes fraction",
fontsize=17,
va="top",
ha="left",
bbox=dict(boxstyle="round,pad=0.1", facecolor="white", edgecolor="k", pad=3.0),
)
ax.set_yticks([0.995, 1])
ax.yaxis.tick_right()
ax.yaxis.set_major_formatter(mpl.ticker.FormatStrFormatter("%.3f"))
_fig.tight_layout()
_fig.savefig(
f"../paper/figures/Ross_Sea_0{i}_eq_sources.png", dpi=300, bbox_inches="tight"
)
number value datetime_start datetime_complete \
0 0 0.648140 2025-10-14 17:26:24.563757 2025-10-14 17:26:26.896538
2 2 -4.806474 2025-10-14 17:26:30.282361 2025-10-14 17:26:33.541067
4 4 0.998421 2025-10-14 17:26:36.483900 2025-10-14 17:26:38.938787
15 15 0.998077 2025-10-14 17:27:05.769515 2025-10-14 17:27:08.028613
9 9 0.998313 2025-10-14 17:26:53.209184 2025-10-14 17:26:54.152648
18 18 0.997754 2025-10-14 17:27:11.323140 2025-10-14 17:27:12.240750
5 5 0.998274 2025-10-14 17:26:38.956968 2025-10-14 17:26:40.007788
17 17 0.998367 2025-10-14 17:27:10.209005 2025-10-14 17:27:11.320529
10 10 0.997820 2025-10-14 17:26:54.156784 2025-10-14 17:26:55.466887
16 16 0.997945 2025-10-14 17:27:08.043038 2025-10-14 17:27:10.206136
3 3 0.997784 2025-10-14 17:26:33.575002 2025-10-14 17:26:36.482301
12 12 0.997467 2025-10-14 17:26:57.375020 2025-10-14 17:26:59.096559
7 7 0.996842 2025-10-14 17:26:45.989873 2025-10-14 17:26:50.215588
14 14 0.996549 2025-10-14 17:27:02.027629 2025-10-14 17:27:05.767862
6 6 0.996640 2025-10-14 17:26:40.009620 2025-10-14 17:26:45.987381
11 11 0.995851 2025-10-14 17:26:55.491321 2025-10-14 17:26:57.371126
19 19 0.995826 2025-10-14 17:27:12.242155 2025-10-14 17:27:14.045315
8 8 0.995811 2025-10-14 17:26:50.216991 2025-10-14 17:26:53.192063
13 13 0.995632 2025-10-14 17:26:59.099337 2025-10-14 17:27:02.025740
1 1 0.994915 2025-10-14 17:26:26.898516 2025-10-14 17:26:30.262121
duration params_depth system_attrs_fixed_params state
0 0 days 00:00:02.332781 100.000000 {'depth': 100} COMPLETE
2 0 days 00:00:03.258706 59563.155753 NaN COMPLETE
4 0 days 00:00:02.454887 209538.155753 NaN COMPLETE
15 0 days 00:00:02.259098 220330.827789 NaN COMPLETE
9 0 days 00:00:00.943464 235979.636210 NaN COMPLETE
18 0 days 00:00:00.917610 254022.362720 NaN COMPLETE
5 0 days 00:00:01.050820 272567.639566 NaN COMPLETE
17 0 days 00:00:01.111524 293970.714442 NaN COMPLETE
10 0 days 00:00:01.310103 316621.779683 NaN COMPLETE
16 0 days 00:00:02.163098 337234.838605 NaN COMPLETE
3 0 days 00:00:02.907299 359513.155753 NaN COMPLETE
12 0 days 00:00:01.721539 390544.833609 NaN COMPLETE
7 0 days 00:00:04.225715 420429.453363 NaN COMPLETE
14 0 days 00:00:03.740233 451337.710587 NaN COMPLETE
6 0 days 00:00:05.977761 482052.792374 NaN COMPLETE
11 0 days 00:00:01.879805 513572.232574 NaN COMPLETE
19 0 days 00:00:01.803160 529463.355119 NaN COMPLETE
8 0 days 00:00:02.975072 545339.385032 NaN COMPLETE
13 0 days 00:00:02.926403 576328.090635 NaN COMPLETE
1 0 days 00:00:03.363605 600000.000000 {'depth': 600000.0} COMPLETE
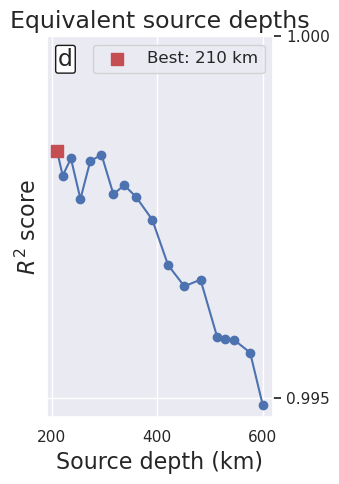
[73]:
fig.image(
imagefile=f"../paper/figures/Ross_Sea_0{i}_eq_sources.png",
position="n1.01/.48+jML",
)
fig.savefig(f"../paper/figures/Ross_Sea_0{i}_grav_anomalies.png")
fig.show()
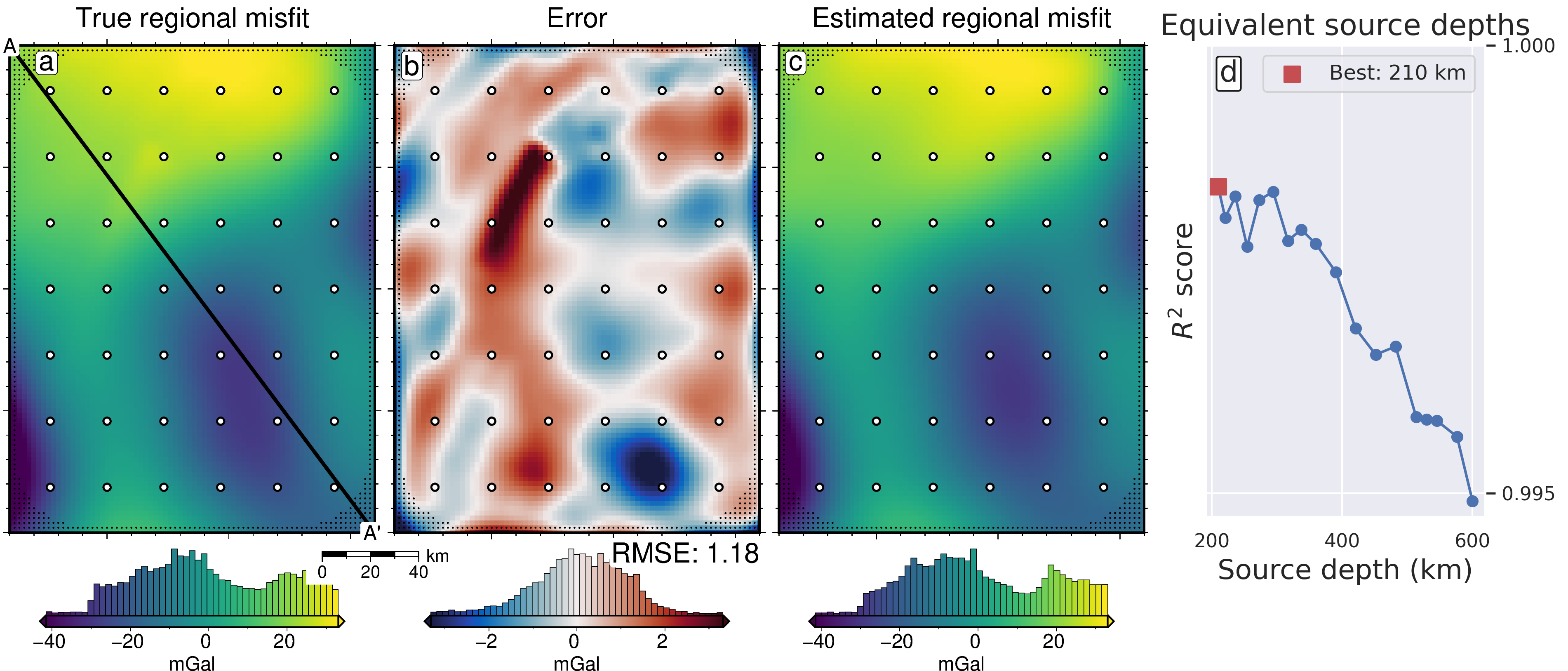
[29]:
grav_grid = grav_df.set_index(["northing", "easting"]).to_xarray()
grav_grid["true_reg"] = grav_grid.basement_grav
titles = [
"misfit",
"residual",
"regional",
"true regional",
]
grids = [
grav_grid.misfit,
grav_grid.res,
grav_grid.reg,
grav_grid.true_reg,
]
data_ylims = None
data_dict = profiles.make_data_dict(
names=titles,
grids=grids,
colors=[ # https://www.simplifiedsciencepublishing.com/resources/best-color-palettes-for-scientific-figures-and-data-visualizations
"216/48/52", # red, d83034
"0/58/125", # dark blue, 003a7d
"78/203/141", # green, 4ecb8d
f"{78 * 0.7}/{203 * 0.7}/{141 * 0.7}", # green, darker than above
# "199/1/255", # purple, c701ff
# "255/157/58", # orange, ff9d3a
# "0/141/255", # med blue, 008dff
# "255/115/182", # pink, ff73b6
# "249/232/88", # yellow, f9e858
],
)
fig, df_data = profiles.plot_data(
"points",
start=profile_start,
stop=profile_end,
num=10000,
fig_height=4,
fig_width=14,
data_dict=data_dict,
data_legend_loc="jTR+o0c/-.2c",
data_buffer=0.01,
data_frame=["neSW", "xafg+lDistance (m)", "yag+lUncertainty (m)"],
data_pen_style=[None, None, None, "4_2:2p"],
data_pen_thickness=[2, 2, 2, 1.5],
share_yaxis=True,
start_label="A",
end_label="A' ",
)
# get grid of min dists to nearest constraints
min_dist = utils.normalized_mindist(
constraint_points,
grids[0],
)
# set y height in data units for add dist to constraint points
data_max = df_data.drop(columns=["easting", "northing", "dist"]).max().max()
if data_ylims is not None:
data_max = data_ylims[1]
df_data = profiles.sample_grids(
df_data, min_dist, interpolation="b", sampled_name="min_dist"
)
# set color limits
series = [1e3, 10e3]
pygmt.makecpt(
cmap="gray",
series=series,
background=True,
)
fig.shift_origin(
yshift=".2c",
)
fig.plot(
x=df_data.dist,
y=np.ones_like(df_data.dist) * data_max,
style="c0.1c",
fill=df_data.min_dist,
cmap=True,
no_clip=True,
)
fig.shift_origin(
yshift="-.2c",
)
with pygmt.config(
FONT="26p,Helvetica,black",
):
fig.colorbar(
cmap=True,
position="jMR+w2c/.3c+jML+v+o0.2c/0c+e",
frame=["xa+lnearest constraint", "y+lkm"],
scale=0.001,
)
fig.savefig(f"../paper/figures/Ross_Sea_0{i}_grav_anomalies_profiles.png")
fig.show()
grdtrack [WARNING]: Some input points were outside the grid domain(s).
grdtrack [WARNING]: Some input points were outside the grid domain(s).
grdtrack [WARNING]: Some input points were outside the grid domain(s).
grdtrack [WARNING]: Some input points were outside the grid domain(s).
grdtrack [WARNING]: Some input points were outside the grid domain(s).
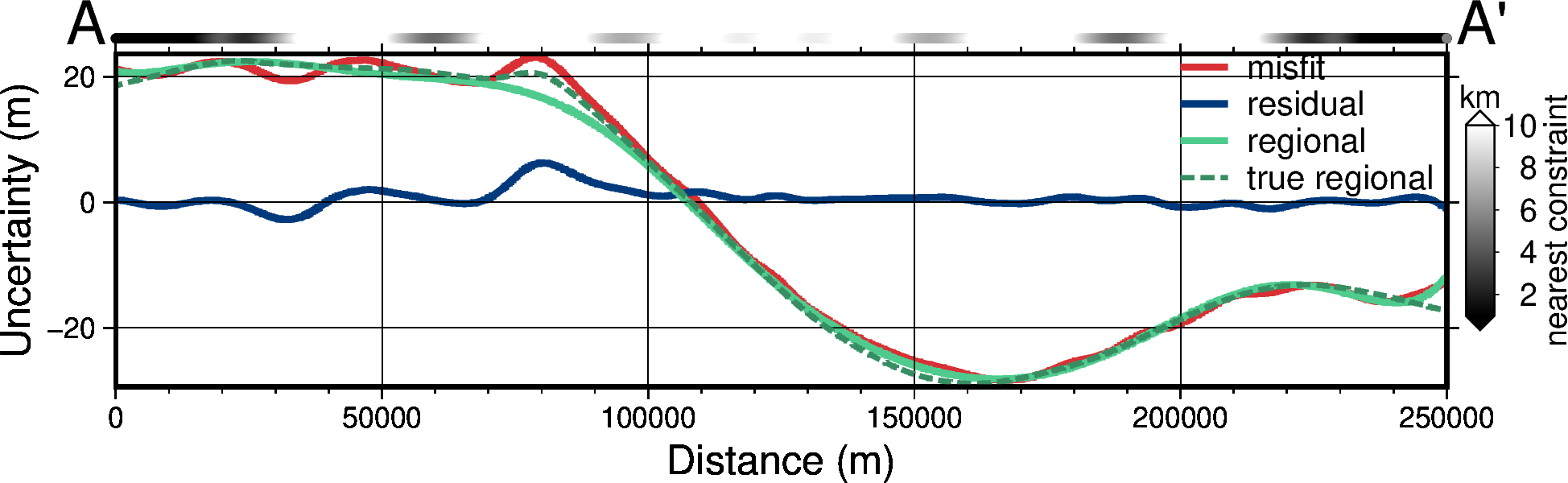
[75]:
grav_error = np.abs(grav_df.gravity_anomaly_full_res_no_noise - grav_df.gravity_anomaly)
grav_error.describe()
[75]:
count 7676.000000
mean 0.847248
std 0.755891
min 0.000369
25% 0.302118
50% 0.651053
75% 1.179984
max 8.188145
dtype: float64
[76]:
reg_error = np.abs(grav_df.basement_grav - grav_df.reg)
reg_error.describe()
[76]:
count 7676.000000
mean 0.897273
std 0.770416
min 0.000056
25% 0.332068
50% 0.721497
75% 1.241281
max 7.489381
dtype: float64
[77]:
polar_utils.get_min_max(np.abs(final_bathymetry - bathymetry))
[77]:
(np.float64(0.004187297208432028), np.float64(109.49392183017909))
[30]:
titles = [
"True error",
"Total uncertainty",
"Uncert. from damping",
"Uncert. from density",
"Uncert. from gravity",
"Uncert. from interpolation",
]
grids = [
np.abs(final_bathymetry - bathymetry),
sensitivity_ds.full_stdev,
sensitivity_ds.damping_stdev,
sensitivity_ds.density_stdev,
sensitivity_ds.grav_stdev,
sensitivity_ds.eq_sources_stdev,
]
sensitivity_plot(
titles,
grids,
constraint_points=constraint_points,
num_in_top_row=3,
robust=False,
fname=f"../paper/figures/Ross_Sea_0{i}_sensitivity.png",
)
gmtset [WARNING]: Representation of font type not recognized. Using default.
gmtset [WARNING]: Representation of font type not recognized. Using default.
gmtset [WARNING]: Representation of font type not recognized. Using default.
gmtset [WARNING]: Representation of font type not recognized. Using default.
gmtset [WARNING]: Representation of font type not recognized. Using default.
gmtset [WARNING]: Representation of font type not recognized. Using default.
[30]:
All together¶
Damping CV’s¶
[ ]:
fig_c, ax = plt.subplots(figsize=(5, 3.5))
stdevs = [
0.24436989744708637, # test 1
0.23960638100341206, # test 2
0.24466075491969516, # test 3
0.2412652540738441, # test 4
0.23771691803388237, # test 5
]
for i, j in enumerate(damping_studies):
scores = j.trials_dataframe().value.to_numpy()
parameters = j.trials_dataframe().params_damping.to_numpy()
param_name = "Damping"
df0 = pd.DataFrame({"scores": scores, "parameters": parameters})
df = df0.sort_values(by="parameters")
best = df.scores.argmin()
color = sns.color_palette("tab10")[i]
ax.plot(
df.parameters,
df.scores,
color=color,
)
# plot +/- stdev's
best_damp = float(df.parameters.iloc[best])
upper = float(10 ** (np.log10(best_damp) + stdevs[i]))
lower = float(10 ** (np.log10(best_damp) - stdevs[i]))
print(best_damp)
# plot square for best value
ax.plot(
df.parameters.iloc[best],
df.scores.iloc[best],
"s",
markersize=10,
color=color,
label=f"Test {i + 1}) {inversion_titles[i]}",
zorder=2,
)
# plot lines for plus and minus stdev
ax.plot(
lower,
df.scores.iloc[best],
"|",
markersize=20,
color=color,
markeredgewidth=1.5,
zorder=40,
)
ax.plot(
upper,
df.scores.iloc[best],
"|",
markersize=20,
color=color,
markeredgewidth=1.5,
zorder=40,
)
ax.set_xscale("log")
ax.set_yscale("log")
ax.set_xlabel("Damping value")
ax.set_ylabel("Trial score")
ax.set_title("c) Damping cross-validation")
plt.tight_layout()
# plt.savefig(
# "../paper/figures/Ross_Sea_damping_cvs.png",
# dpi=300,
# )
0.014002832326761543
0.01406058612011891
0.014177009401440639
0.013382994085308205
0.013422413235354692
Density optimizations¶
[ ]:
fig_b, ax = plt.subplots(figsize=(5, 3.5))
stdevs = [
5, # test 1
5, # test 2
20, # test 3
500, # test 4
500, # test 5
]
for i, j in enumerate(density_studies):
scores = j.trials_dataframe().value.to_numpy()
parameters = j.trials_dataframe().params_density_contrast.to_numpy()
param_name = "Density"
df0 = pd.DataFrame({"scores": scores, "parameters": parameters})
df = df0.sort_values(by="parameters")
best = df.scores.argmin()
color = sns.color_palette("tab10")[i]
ax.plot(
df.parameters,
df.scores,
color=color,
)
# plot square for best value
ax.plot(
df.parameters.iloc[best],
df.scores.iloc[best],
"s",
markersize=10,
color=color,
zorder=2,
)
# plot +/- stdev's
upper = df.parameters.iloc[best] + stdevs[i]
lower = df.parameters.iloc[best] - stdevs[i]
# plot lines for plus and minus stdev
ax.plot(
lower,
df.scores.iloc[best],
"|",
markersize=20,
color=color,
markeredgewidth=1.5,
zorder=40,
)
ax.plot(
upper,
df.scores.iloc[best],
"|",
markersize=20,
color=color,
markeredgewidth=1.5,
zorder=40,
)
# plot K-Folds approach for Tests 4 and 5
if i in [3, 4]:
f = f"../results/Ross_Sea/Ross_Sea_0{i + 1}_density_cv_study.pickle"
with pathlib.Path(f).open("rb") as f:
r = pickle.load(f)
scores = r.trials_dataframe().value.to_numpy()
parameters = r.trials_dataframe().params_density_contrast.to_numpy()
param_name = "Density"
df0 = pd.DataFrame({"scores": scores, "parameters": parameters})
df = df0.sort_values(by="parameters")
best = df.scores.argmin()
color = sns.color_palette("tab10")[i]
ax.plot(
df.parameters,
df.scores,
color=color,
linestyle="dotted",
)
# plot square for best value
ax.plot(
df.parameters.iloc[best],
df.scores.iloc[best],
"s",
markersize=6,
color=color,
zorder=2,
)
y_lims = ax.get_ylim()
ax.vlines(
true_density_contrast,
ymin=y_lims[0],
ymax=y_lims[1],
color="k",
zorder=1,
label="True density",
)
ax.set_xlabel("Density contrast (kg/m³)")
ax.set_ylabel("Trial score")
ax.set_title("d) Density contrast estimation")
plt.yscale("log")
plt.tight_layout()
# plt.savefig(
# "../paper/figures/Ross_Sea_density_optimizations.png",
# dpi=300,
# )
[ ]:
fig, ax1 = plt.subplots(figsize=(5, 3.5))
for i, j in enumerate(optimal_results):
grav_results = j[1]
params = j[2]
# get misfit data at end of each iteration
cols = [s for s in grav_results.columns.to_list() if "_final_misfit" in s]
iters = len(cols)
l2_norms = [np.sqrt(utils.rmse(grav_results[i])) for i in cols]
starting_misfit = utils.rmse(grav_results["iter_1_initial_misfit"])
starting_l2_norm = np.sqrt(starting_misfit)
# add starting l2 norm to the beginning of the list
l2_norms.insert(0, starting_l2_norm)
# calculate delta L2-norms
delta_l2_norms = []
for k, m in enumerate(l2_norms):
if k == 0:
delta_l2_norms.append(np.nan)
else:
delta_l2_norms.append(l2_norms[k - 1] / m)
# get tolerance values
l2_norm_tolerance = float(params["L2 norm tolerance"])
delta_l2_norm_tolerance = float(params["Delta L2 norm tolerance"])
color = sns.color_palette("tab10")[i]
# plot L2-norm convergence
ax1.plot(
range(iters + 1),
l2_norms,
color=color,
)
# plot square for end iteration
ax1.plot(
iters,
l2_norms[-1],
"s",
markersize=8,
color=color,
zorder=2,
label=f"Test {i + 1}) {inversion_titles[i]}",
)
# set x axis to integer values
ax1.xaxis.set_major_locator(mpl.ticker.MaxNLocator(integer=True))
ax1.legend(loc="upper right", fontsize=10)
# set axis labels, ticks and gridlines
ax1.set_xlabel("Iteration")
ax1.set_ylabel("L2-norm")
ax1.tick_params(axis="y", which="both")
plt.title("d) Inversion convergence")
plt.tight_layout()
# plt.savefig(
# "../paper/figures/Ross_Sea_convergences.png",
# dpi=300,
# )
plt.show()
Power spectrum¶
[82]:
dif_grids = []
for g in final_bathymetries:
# for g in final_stochastic_means:
dif_grids.append(bathymetry - g)
# set min max as 2nd to last grid
dif_lims = polar_utils.get_min_max(dif_grids[-2], robust=True)
dif_lims
[82]:
(np.float64(-21.621033707139702), np.float64(12.798915128951933))
[83]:
names = ["true", 5, 10, 20]
true = bathymetry
grids = [
true,
*[
inv_synthetic.contaminate_with_long_wavelength_noise(
true,
coarsen_factor=None,
spacing=x * 1e3,
noise_as_percent=False,
noise=50,
)
for x in names[1:]
],
]
fig = maps.subplots(
grids,
hist=True,
cmap="viridis",
scalebar=True,
)
fig.show()
fig = maps.subplots(
[g - true for g in grids],
hist=True,
cmap="balance+h0",
scalebar=True,
)
fig.show()
requested spacing (2000.0) is smaller than the original (5000.0).
requested spacing (2000.0) is smaller than the original (10000.0).
requested spacing (2000.0) is smaller than the original (20000.0).
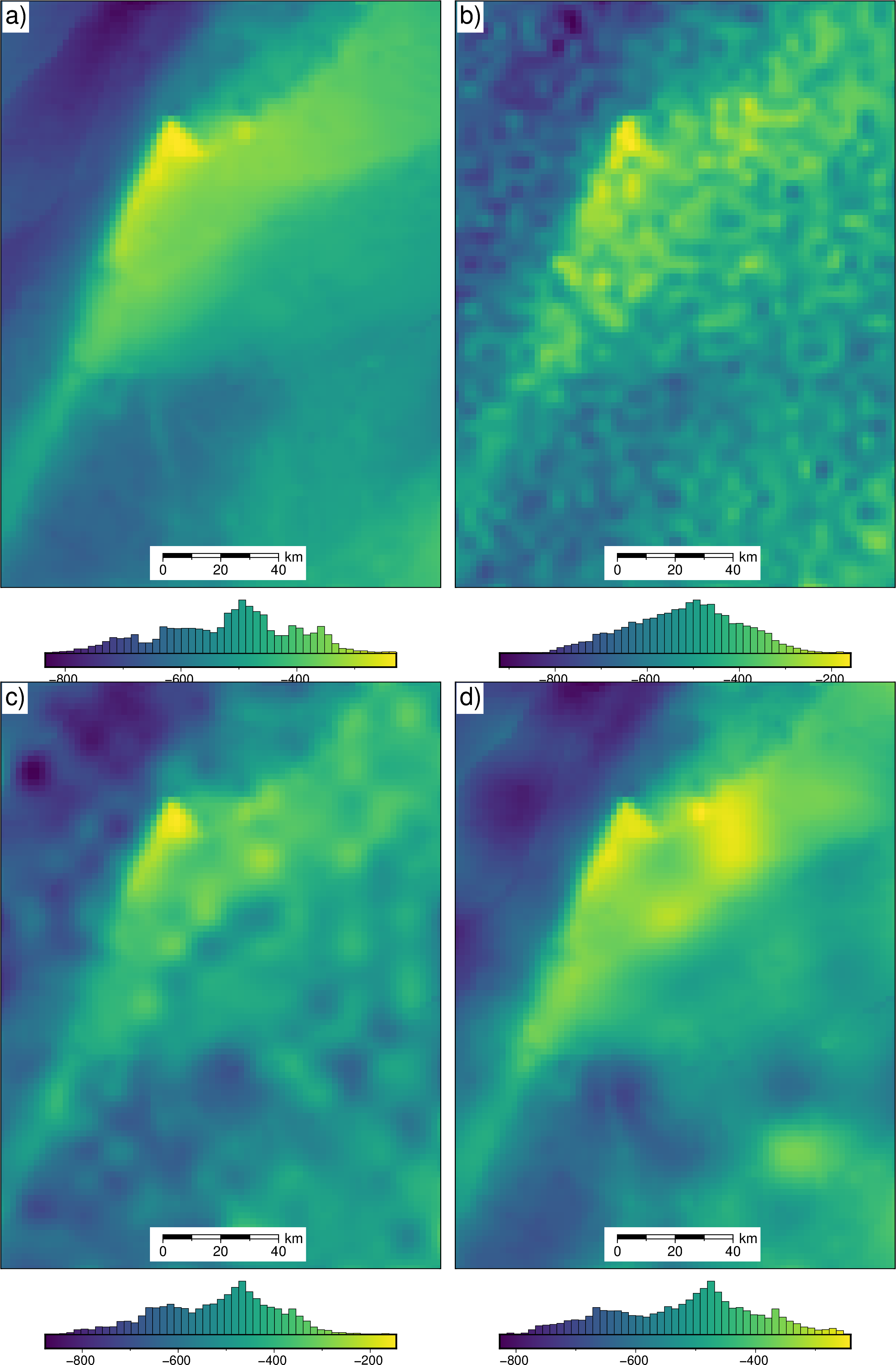
makecpt [ERROR]: Option T: min >= max
supplied min value is greater or equal to max value
Grid/points are a constant value, can't make a colorbar histogram!
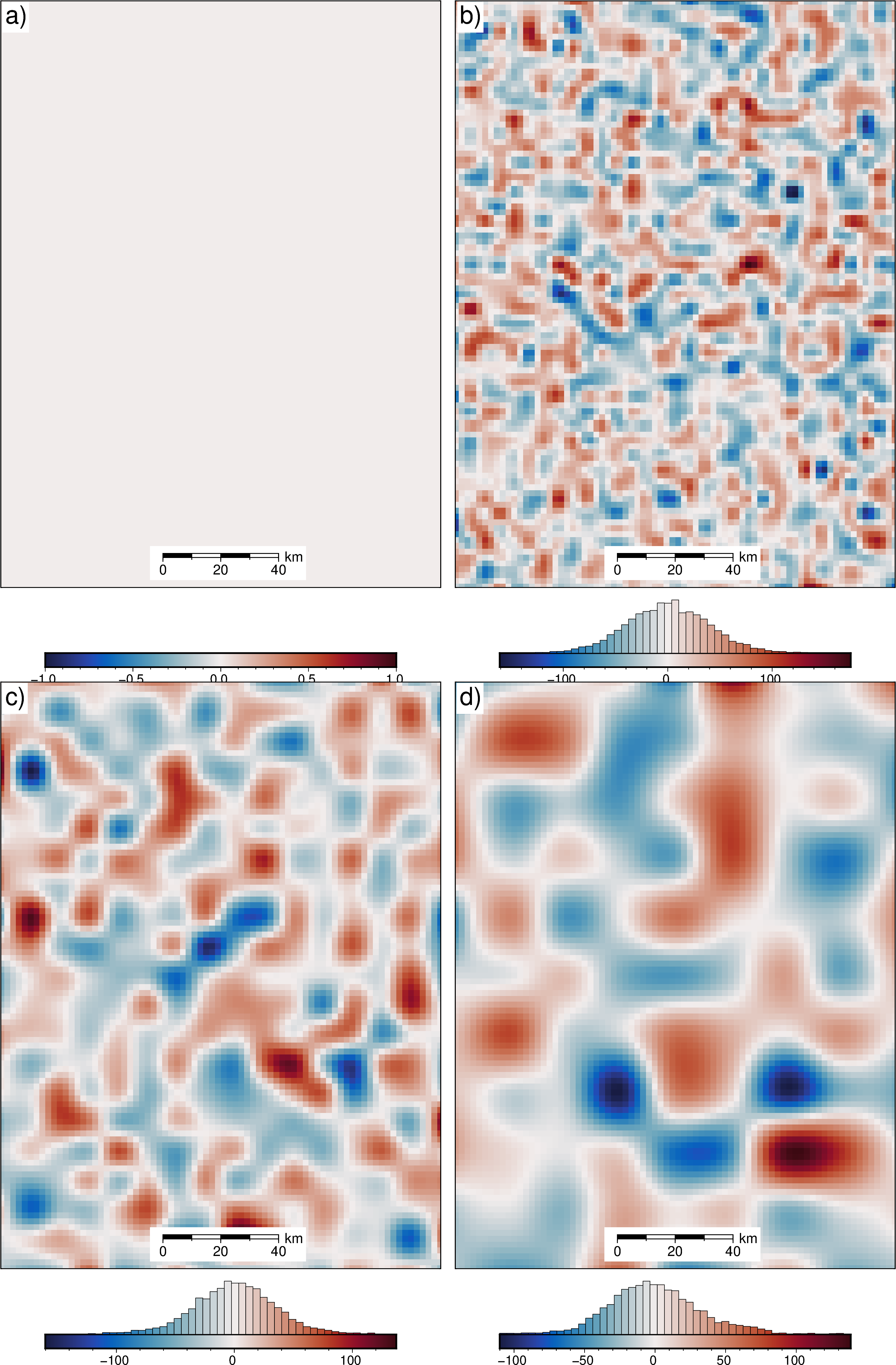
[84]:
names = [
"Starting bathymetry",
*[inversion_titles[i] for i in range(len(dif_grids))],
]
grids = [
bathymetry - starting_bathymetry,
*dif_grids,
]
raps_das = []
for i, g in enumerate(grids):
raps_das.append(
xrft.isotropic_power_spectrum(
g,
dim=["easting", "northing"],
detrend="linear",
truncate=False,
)
)
fig_b, ax = plt.subplots(figsize=(5, 3.5))
for i, da in enumerate(raps_das):
c = "black" if i == 0 else sns.color_palette("tab10")[i - 1]
ax.plot(
(1 / da.freq_r) / 1e3,
da,
label=names[i],
color=c,
)
ax.set_xscale("log")
ax.set_yscale("log")
leg = ax.legend(
loc="upper left",
fontsize=9,
labelspacing=0.2,
handlelength=1,
)
for legobj in leg.legend_handles:
legobj.set_linewidth(3)
ax.set_xlabel("Wavelength (km)")
ax.set_ylabel("Power ($m^3$)")
ax.set_title("b) Radially averaged PSD of errors")
plt.tight_layout()
ax.set_xticks(
[
5,
10,
20,
40,
80,
]
)
ax.xaxis.set_major_formatter(mpl.ticker.StrMethodFormatter("{x:.0f}"))
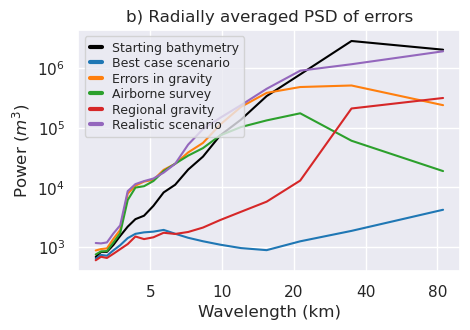
Inversion results¶
[85]:
# use combined min max
inversion_error_lims = (
0,
polar_utils.get_combined_min_max(abs_inversion_errors, robust=True)[1],
)
# set min max as 2nd to last grid
uncert_error_lims = polar_utils.get_min_max(uncert_errors[-2], robust=True)
inversion_error_lims, uncert_error_lims
[85]:
((0, np.float64(58.26295740916967)),
(np.float64(-7.316209640741482), np.float64(17.239395411739565)))
[110]:
inversion_MAEs = []
inversion_RMSEs = []
uncert_MAEs = []
uncert_RMSEs = []
uncert_MAs = []
uncert_RMSs = []
constraint_points = constraint_points_dfs[0]
titles = [*inversion_titles, "Starting bathymetry"]
# set cmap lims
bathy_lims = polar_utils.get_min_max(
bathymetry,
)
bathy_error_lims = (
0,
polar_utils.get_combined_min_max(
np.abs([*dif_grids, bathymetry - starting_bathymetry]),
robust=True,
robust_percentiles=(0.07, 0.93),
)[1],
)
uncert_lims = (
0,
polar_utils.get_combined_min_max(
final_uncertainties,
robust=True,
robust_percentiles=(0.005, 0.995),
)[1],
)
bathy_error_lims = (0, 40)
uncert_lims = bathy_error_lims
grds = [*final_bathymetries, starting_bathymetry]
for i, grd in enumerate(grds):
if i == 0:
fig = None
origin_shift = "initialize"
scalebar = True
else:
origin_shift = "both_shift"
scalebar = False
if i == len(grds) - 1:
uncert_grd = None
bathy_dif = bathymetry - grd
else:
uncert_grd = final_uncertainties[i]
bathy_dif = dif_grids[i]
# plot the inverted bathymetry
fig = maps.plot_grd(
grd,
fig_height=10,
cmap="rain",
reverse_cpt=True,
cpt_lims=bathy_lims,
cbar_unit="m",
cbar_font="28p,Helvetica,black",
frame=["nswe", "xaf10000", "yaf10000"],
hist=True,
hist_bin_num=35,
fig=fig,
origin_shift=origin_shift,
yshift_amount=2.1,
points=constraint_points[~constraint_points.inside],
points_style="p1p",
scalebar=scalebar,
scalebar_position="jBL+o-.5c/-.6c",
)
fig.plot(
x=constraint_points[constraint_points.inside].easting,
y=constraint_points[constraint_points.inside].northing,
style="c.15c",
fill="white",
pen="1p,black",
)
# add title to each column
fig.text(
position="TC",
justify="BC",
text=titles[i],
fill="white",
font="25p,Helvetica,black",
clearance="+tO",
offset="j0/.8",
no_clip=True,
)
if i == len(grds) - 1:
pass
else:
fig.text(
position="TC",
justify="BC",
text=f"Test {i + 1}",
fill="white",
font="25p,Helvetica,black",
clearance="+tO",
offset="j0/1.9",
no_clip=True,
)
if i == 0:
# add title to top row
fig.text(
position="ML",
justify="BC",
offset="j0/1",
text="Inverted bathymetry",
fill="white",
angle=90,
font="26p,Helvetica,black",
clearance="+tO",
no_clip=True,
)
# plot profiles location, and endpoints on map
fig.plot(
vd.line_coordinates(profile_start, profile_end, size=100),
pen="2p,black",
)
fig.text(
x=profile_start[0],
y=profile_start[1],
text="A",
fill="white",
font="20p,Helvetica,black",
justify="BR",
clearance="+tO",
no_clip=True,
)
fig.text(
x=profile_end[0],
y=profile_end[1],
text="A' ",
fill="white",
font="20p,Helvetica,black",
justify="TL",
clearance="+tO",
no_clip=True,
)
# plot subplot labels (1a, 1b, 1c, etc.)
fig.text(
position="TL",
text=f"{i + 1}a",
fill="white",
font="20p,Helvetica,black",
offset="j.2/.2",
clearance="+tO",
)
inversion_MAE = np.abs(bathy_dif).to_numpy().mean()
inversion_RMSE = utils.rmse(bathy_dif)
# add to lists
inversion_MAEs.append(inversion_MAE)
inversion_RMSEs.append(inversion_RMSE)
# plot bathymetry error
fig = maps.plot_grd(
np.abs(bathy_dif),
fig_height=10,
cmap="ice",
reverse_cpt=True,
cpt_lims=bathy_error_lims,
cbar_unit="m",
cbar_font="28p,Helvetica,black",
frame=["nswe", "xaf10000", "yaf10000"],
hist=True,
hist_bin_num=35,
fig=fig,
origin_shift="y",
yshift_amount=-1.05,
points=constraint_points[~constraint_points.inside],
points_style="p1p",
)
fig.plot(
x=constraint_points[constraint_points.inside].easting,
y=constraint_points[constraint_points.inside].northing,
style="c.15c",
fill="white",
pen="1p,black",
)
# add errors to plot
fig.text(
position="BR",
justify="TR",
text=f"RMSE: {round(inversion_RMSE, 1)}",
fill="white",
font="18p,Helvetica,black",
offset="j-.5/.2",
clearance="+tO",
no_clip=True,
)
# plot subplot labels
fig.text(
position="TL",
text=f"{i + 1}b",
fill="white",
font="20p,Helvetica,black",
offset="j.2/.2",
clearance="+tO",
)
if i == 0:
# add title to 2nd row
fig.text(
position="ML",
justify="BC",
offset="j0/1",
text="Bathymetry error",
fill="white",
angle=90,
font="26p,Helvetica,black",
clearance="+tO",
no_clip=True,
)
# plot the uncertainty
if i == len(grds) - 1:
uncert_MA = None
uncert_RMS = None
uncert_MAE = None
uncert_RMSE = None
else:
uncert_MA = np.abs(uncert_grd.to_numpy()).mean()
uncert_RMS = utils.rmse(uncert_grd)
uncert_MAE = np.abs(uncert_errors[i]).to_numpy().mean()
uncert_RMSE = utils.rmse(uncert_errors[i])
uncert_MAs.append(uncert_MA)
uncert_RMSs.append(uncert_RMS)
uncert_MAEs.append(uncert_MAE)
uncert_RMSEs.append(uncert_RMSE)
if i == len(grds) - 1:
continue
fig = maps.plot_grd(
uncert_grd,
fig_height=10,
cmap="ice",
reverse_cpt=True,
cpt_lims=uncert_lims,
cbar_unit="m",
cbar_font="28p,Helvetica,black",
frame=["nswe", "xaf10000", "yaf10000"],
hist=True,
hist_bin_num=35,
fig=fig,
origin_shift="y",
yshift_amount=-1.05,
points=constraint_points[~constraint_points.inside],
points_style="p1p",
)
fig.plot(
x=constraint_points[constraint_points.inside].easting,
y=constraint_points[constraint_points.inside].northing,
style="c.15c",
fill="white",
pen="1p,black",
)
# add mean / rms to plot
fig.text(
position="BR",
justify="TR",
text=f"RMS: {round(uncert_RMS, 1)}",
fill="white",
font="18p,Helvetica,black",
offset="j-.5/.2",
clearance="+tO",
no_clip=True,
)
# plot subplot labels
fig.text(
position="TL",
text=f"{i + 1}c",
fill="white",
font="20p,Helvetica,black",
offset="j.2/.2",
clearance="+tO",
)
if i == 0:
# add title to 3rd row
fig.text(
position="ML",
justify="BC",
offset="j0/1",
text="Estimated uncertainty",
fill="white",
angle=90,
font="26p,Helvetica,black",
clearance="+tO",
no_clip=True,
)
fig.savefig("../paper/figures/Ross_Sea_inversions.png", dpi=500)
fig.show()
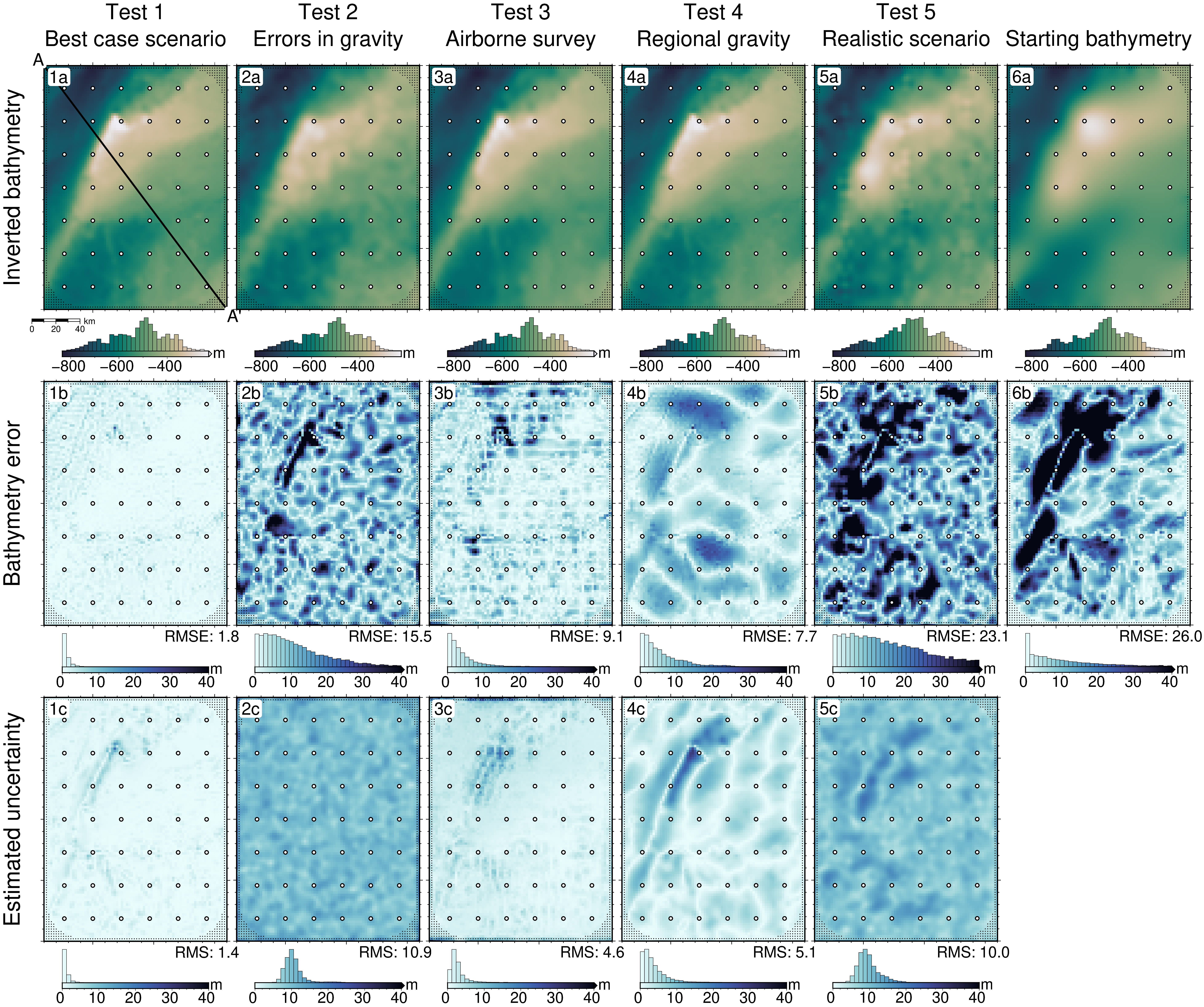
Sensitivity results¶
Inversion result plots¶
[7]:
hexs = sns.color_palette("tab10").as_hex()[:]
rgbs = [PIL.ImageColor.getrgb(h) for h in hexs]
colors = [f"{r[0]}/{r[1]}/{r[2]}" for r in rgbs]
[8]:
inversion_titles
[8]:
['Best case scenario',
'Errors in gravity',
'Airborne survey',
'Regional gravity',
'Realistic scenario']
[89]:
titles = [
"Test 1",
"Test 2",
"Test 3",
"Test 4",
"Test 5",
"Starting\nbathymetry",
]
df = pd.DataFrame(
{
"inversion_RMSE": inversion_RMSEs,
"inversion_MAE": inversion_MAEs,
"title": titles,
"color": sns.color_palette("tab10")[0 : len(inversion_RMSEs) - 1] + ["black"], # noqa: RUF005
}
)
fig_a, ax1 = plt.subplots(figsize=(5, 3.5))
ax1.scatter(
data=df,
x="title",
y="inversion_RMSE",
color=df.color,
marker="^",
s=100,
edgecolor="k",
)
ax1.set_ylabel("meters")
plt.title("Inversion errors")
inversion_label = mpl.lines.Line2D(
[0],
[0],
label="Bathymetry RMSE",
marker="^",
markersize=10,
markeredgecolor="k",
markerfacecolor="none",
linestyle="",
)
plt.legend(handles=[inversion_label])
plt.tight_layout()
plt.show()
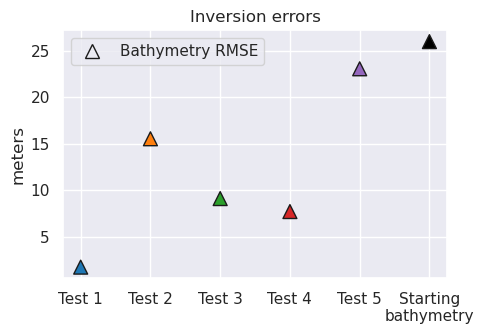
[ ]:
titles = [
"Test 1",
"Test 2",
"Test 3",
"Test 4",
"Test 5",
"Starting\nbathymetry",
]
df = pd.DataFrame(
{
"inversion_RMSE": inversion_RMSEs,
"uncertainty_RMSE": uncert_RMSEs,
"inversion_MAE": inversion_MAEs,
"uncertainty_MAE": uncert_MAEs,
"uncertainty_MAs": uncert_MAs,
"uncertainty_RMSs": uncert_RMSs,
"title": titles,
"color": sns.color_palette("tab10")[0 : len(inversion_RMSEs) - 1] + ["black"], # noqa: RUF005
}
)
fig_a, ax1 = plt.subplots(figsize=(5, 3.5))
ax1.scatter(
data=df,
x="title",
y="inversion_RMSE",
color=df.color,
marker="^",
s=100,
edgecolor="k",
)
ax1.scatter(
data=df,
x="title",
y="uncertainty_RMSs",
color=df.color,
marker="s",
s=80,
edgecolor="k",
)
ax1.set_ylabel("meters")
plt.title("a) Errors and uncertainties")
inversion_label = mpl.lines.Line2D(
[0],
[0],
label="Bathymetry RMSE",
marker="^",
markersize=10,
markeredgecolor="k",
markerfacecolor="none",
linestyle="",
)
uncert_mean_label = mpl.lines.Line2D(
[0],
[0],
label="Uncertainty RMS",
marker="s",
markersize=10,
markeredgecolor="k",
markerfacecolor="none",
linestyle="",
)
plt.legend(handles=[inversion_label, uncert_mean_label])
plt.tight_layout()
# plt.savefig(
# "../paper/figures/Ross_Sea_inversion_errors.png",
# dpi=300,
# )
plt.show()
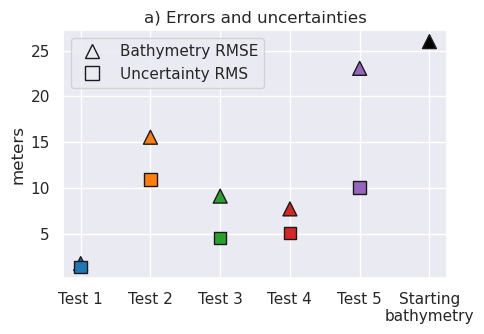
[91]:
df.inversion_RMSE - df.uncertainty_RMSs
[91]:
0 0.385471
1 4.629030
2 4.555937
3 2.656253
4 13.019786
5 NaN
dtype: float64
[29]:
len(final_bathymetries)
[29]:
5
[14]:
[
"black",
*colors[0 : len(inversion_titles)],
"black",
]
[14]:
['black',
'31/119/180',
'255/127/14',
'44/160/44',
'214/39/40',
'148/103/189',
'black']
[13]:
# topography dictionary
topo_names = ["Starting bed", *inversion_titles, "True bed"]
topo_grids = [starting_bathymetry, *final_bathymetries, bathymetry]
layers_dict = profiles.make_data_dict(
names=topo_names,
grids=topo_grids,
colors=[
"black",
*colors[0 : len(inversion_titles)],
"black",
],
)
fig, df_layers, df_data = profiles.plot_profile(
"points",
start=profile_start,
stop=profile_end,
num=100000,
fig_height=5.5,
fig_width=18,
share_yaxis=True,
layers_dict=layers_dict,
layer_buffer=0.03,
layers_legend_loc="jBR+jBR+o.05c/.03c+w9c",
layers_legend_box=["+gwhite"],
layers_legend_columns=2,
layers_legend_font="12p,Helvetica,black",
fill_layers=False,
layers_frame=["neSW", "xafg+lDistance (m)", "yag+lElevation (m)"],
layers_pen_thickness=[1.2, *[1.2] * len(inversion_titles), 0.5],
layers_pen_style=["-", *[None] * len(inversion_titles), None],
start_label="A",
end_label="A' ",
start_end_pen=None,
start_label_justify="TR",
end_label_justify="TL",
start_end_fill=None,
)
fig.savefig("../paper/figures/Ross_Sea_inversion_profiles.png", dpi=500)
fig.show()
grdtrack [WARNING]: Some input points were outside the grid domain(s).
grdtrack [WARNING]: Some input points were outside the grid domain(s).
grdtrack [WARNING]: Some input points were outside the grid domain(s).
grdtrack [WARNING]: Some input points were outside the grid domain(s).
grdtrack [WARNING]: Some input points were outside the grid domain(s).
grdtrack [WARNING]: Some input points were outside the grid domain(s).
grdtrack [WARNING]: Some input points were outside the grid domain(s).
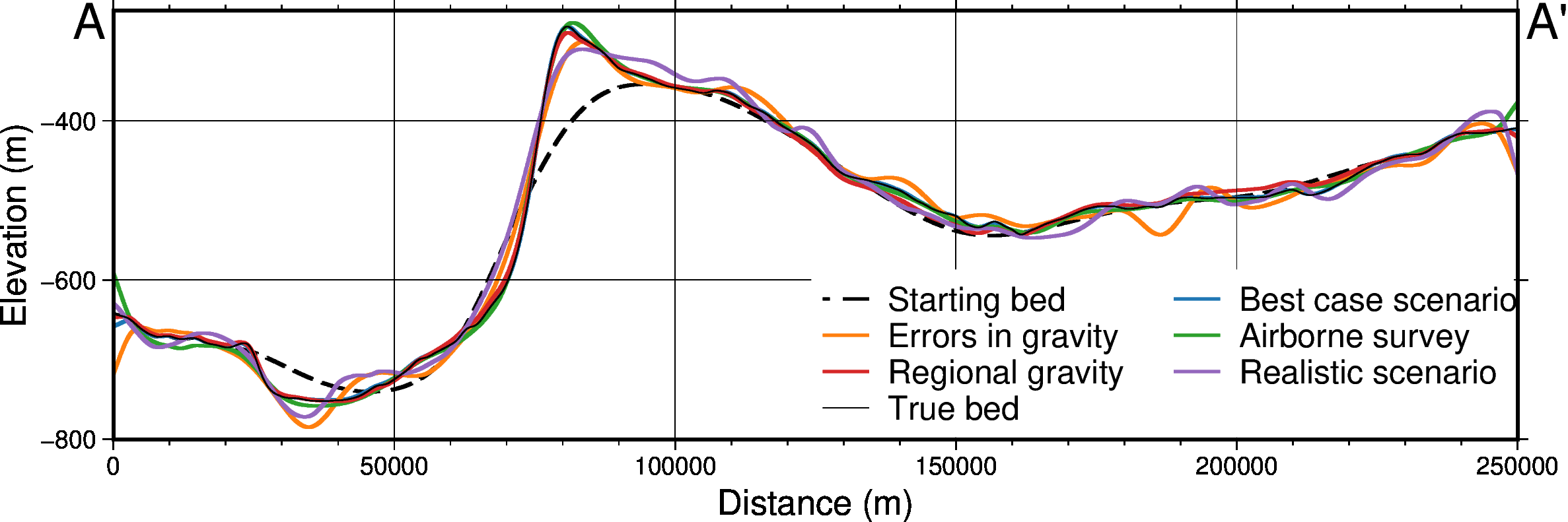
[ ]:
# topography dictionary
topo_names = inversion_titles[::-1]
topo_grids = [x - bathymetry for x in final_bathymetries][::-1]
layers_dict = profiles.make_data_dict(
names=topo_names,
grids=topo_grids,
colors=[
*colors[0 : len(topo_grids)][::-1],
],
)
fig, df_layers, df_data = profiles.plot_profile(
"points",
start=profile_start,
stop=profile_end,
num=100000,
fig_height=4,
fig_width=15,
share_yaxis=True,
layers_dict=layers_dict,
layer_buffer=0.05,
layers_legend_loc="jTR+jTL+o0c/0c",
layers_legend_box=None,
fill_layers=False,
layers_frame=["neSW", "xafg+lDistance (m)", "yag+lError (m)"],
layers_pen_thickness=[1.2] * len(inversion_titles),
start_label="A",
end_label="A' ",
start_end_pen=None,
)
fig.show()
grdtrack [WARNING]: Some input points were outside the grid domain(s).
grdtrack [WARNING]: Some input points were outside the grid domain(s).
grdtrack [WARNING]: Some input points were outside the grid domain(s).
grdtrack [WARNING]: Some input points were outside the grid domain(s).
grdtrack [WARNING]: Some input points were outside the grid domain(s).
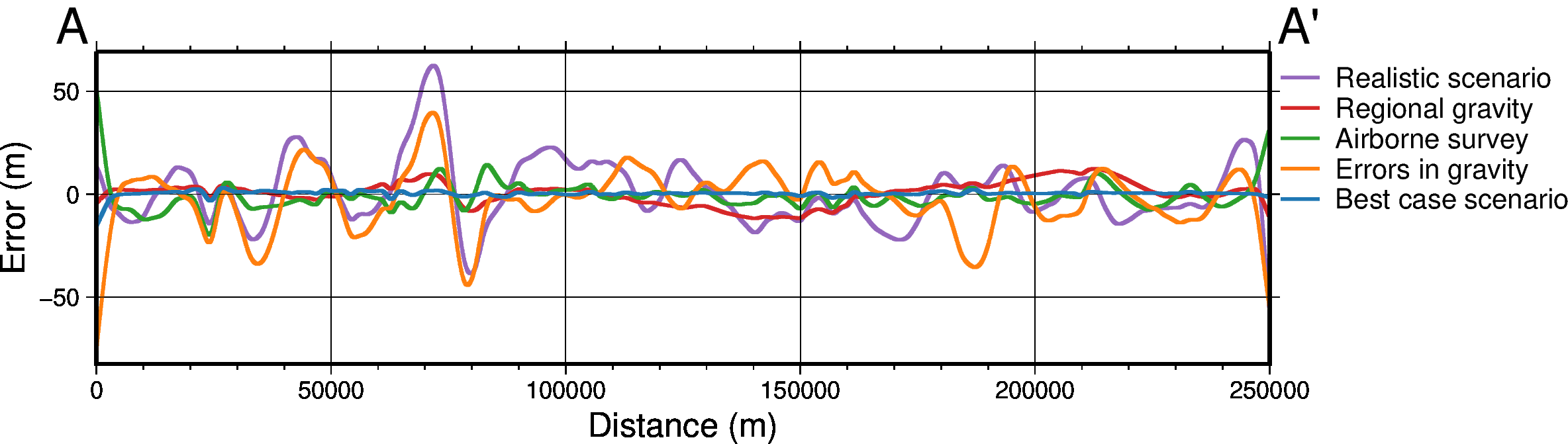
Inversion results subplot¶
[95]:
fig, ((ax1, ax2), (ax3, ax4)) = plt.subplots(
2,
2,
figsize=(5 * 2, 3.5 * 2),
)
###
###
# Errors and uncertainties
###
###
titles = [
"Test 1",
"Test 2",
"Test 3",
"Test 4",
"Test 5",
"Starting\nbathymetry",
]
df = pd.DataFrame(
{
"inversion_RMSE": inversion_RMSEs,
"uncertainty_RMSE": uncert_RMSEs,
"inversion_MAE": inversion_MAEs,
"uncertainty_MAE": uncert_MAEs,
"uncertainty_MAs": uncert_MAs,
"uncertainty_RMSs": uncert_RMSs,
"title": titles,
"color": sns.color_palette("tab10")[0 : len(inversion_RMSEs) - 1] + ["black"], # noqa: RUF005
}
)
ax1.scatter(
data=df,
x="title",
y="inversion_RMSE",
color=df.color,
marker="^",
s=100,
edgecolor="k",
)
ax1.scatter(
data=df,
x="title",
y="uncertainty_RMSs",
color=df.color,
marker="s",
s=80,
edgecolor="k",
)
ax1.set_ylabel("meters")
ax1.set_xticks([])
ax1.set_title("a) Errors and uncertainties", fontsize=16)
inversion_label = mpl.lines.Line2D(
[0],
[0],
label="Bathymetry RMSE",
marker="^",
markersize=10,
markeredgecolor="k",
markerfacecolor="none",
linestyle="",
)
uncert_mean_label = mpl.lines.Line2D(
[0],
[0],
label="Uncertainty RMS",
marker="s",
markersize=10,
markeredgecolor="k",
markerfacecolor="none",
linestyle="",
)
ax1.legend(handles=[inversion_label, uncert_mean_label])
###
###
# Power spectra
###
###
names = [
"Starting bathymetry",
*[f"Test {i + 1}) {inversion_titles[i]}" for i in range(len(dif_grids))],
]
grids = [
bathymetry - starting_bathymetry,
*dif_grids,
]
raps_das = []
for i, g in enumerate(grids):
raps_das.append(
xrft.isotropic_power_spectrum(
g,
dim=["easting", "northing"],
detrend="linear",
)
)
for i, da in enumerate(raps_das):
c = "black" if i == 0 else sns.color_palette("tab10")[i - 1]
lines = ax2.plot(
(1 / da.freq_r) / 1e3,
da,
label=names[i],
color=c,
)
ax2.set_xscale("log")
ax2.set_yscale("log")
leg = fig.legend(
ax2.get_lines(),
[i.get_label() for i in ax2.get_lines()],
loc="lower center",
ncol=3,
bbox_to_anchor=(0.5, -0.08),
fontsize=12,
labelspacing=0.2,
handlelength=0.8,
)
for legobj in leg.legend_handles:
legobj.set_linewidth(6)
ax2.set_xlabel("Wavelength (km)")
ax2.set_ylabel("Power ($m^3$)")
ax2.set_title("b) Radially averaged PSD of errors", fontsize=16)
ax2.set_xticks(
[
# 2,
5,
10,
20,
40,
80,
]
)
ax2.xaxis.set_major_formatter(mpl.ticker.StrMethodFormatter("{x:.0f}"))
###
###
# Damping CV
###
###
stdevs = [
0.2443699101356321, # test 1
0.23929672688818715, # test 2
0.24466078784954876, # test 3
0.24156679945478948, # test 4
0.23934002794701525, # test 5
]
for i, j in enumerate(damping_studies):
scores = j.trials_dataframe().value.to_numpy()
parameters = j.trials_dataframe().params_damping.to_numpy()
param_name = "Damping"
df0 = pd.DataFrame({"scores": scores, "parameters": parameters})
df = df0.sort_values(by="parameters")
best = df.scores.argmin()
color = sns.color_palette("tab10")[i]
ax3.plot(
df.parameters,
df.scores,
color=color,
)
# plot +/- stdev's
best_damp = float(df.parameters.iloc[best])
upper = float(10 ** (np.log10(best_damp) + stdevs[i]))
lower = float(10 ** (np.log10(best_damp) - stdevs[i]))
# plot square for best value
ax3.plot(
df.parameters.iloc[best],
df.scores.iloc[best],
"s",
markersize=10,
color=color,
label=f"Test {i + 1}) {inversion_titles[i]}",
zorder=2,
)
# plot lines for plus and minus stdev
ax3.plot(
lower,
df.scores.iloc[best],
"|",
markersize=20,
color=color,
markeredgewidth=1.5,
zorder=40,
)
ax3.plot(
upper,
df.scores.iloc[best],
"|",
markersize=20,
color=color,
markeredgewidth=1.5,
zorder=40,
)
ax3.set_xscale("log")
ax3.set_yscale("log")
ax3.set_xlabel("Damping value")
ax3.set_ylabel("Trial score")
ax3.set_title("c) Damping cross-validation", fontsize=16)
ax3.yaxis.set_major_formatter(mpl.ticker.StrMethodFormatter("{x:.1f}"))
###
###
# Density
###
###
stdevs = [
5, # test 1
5, # test 2
20, # test 3
500, # test 4
500, # test 5
]
for i, j in enumerate(density_studies):
scores = j.trials_dataframe().value.to_numpy()
parameters = j.trials_dataframe().params_density_contrast.to_numpy()
param_name = "Density"
df0 = pd.DataFrame({"scores": scores, "parameters": parameters})
df = df0.sort_values(by="parameters")
best = df.scores.argmin()
color = sns.color_palette("tab10")[i]
ax4.plot(
df.parameters,
df.scores,
color=color,
)
# plot square for best value
ax4.plot(
df.parameters.iloc[best],
df.scores.iloc[best],
"s",
markersize=10,
color=color,
zorder=2,
)
# plot +/- stdev's
upper = df.parameters.iloc[best] + stdevs[i]
lower = df.parameters.iloc[best] - stdevs[i]
# plot lines for plus and minus stdev
ax4.plot(
lower,
df.scores.iloc[best],
"|",
markersize=20,
color=color,
markeredgewidth=1.5,
zorder=40,
)
ax4.plot(
upper,
df.scores.iloc[best],
"|",
markersize=20,
color=color,
markeredgewidth=1.5,
zorder=40,
)
# plot K-Folds approach for Tests 4 and 5
if i in [3, 4]:
f = f"../results/Ross_Sea/Ross_Sea_0{i + 1}_density_cv_study.pickle"
with pathlib.Path(f).open("rb") as f:
r = pickle.load(f)
scores = r.trials_dataframe().value.to_numpy()
parameters = r.trials_dataframe().params_density_contrast.to_numpy()
param_name = "Density"
df0 = pd.DataFrame({"scores": scores, "parameters": parameters})
df = df0.sort_values(by="parameters")
best = df.scores.argmin()
color = sns.color_palette("tab10")[i]
ax4.plot(
df.parameters,
df.scores,
color=color,
linestyle="dotted",
)
# plot square for best value
ax4.plot(
df.parameters.iloc[best],
df.scores.iloc[best],
"s",
markersize=6,
color=color,
zorder=2,
)
y_lims = ax4.get_ylim()
ax4.vlines(
true_density_contrast,
ymin=y_lims[0],
ymax=y_lims[1],
color="k",
zorder=1,
label="True density",
)
# ax.legend(loc='center left', bbox_to_anchor=(1, 0.5))
ax4.set_xlabel("Density contrast (kg/m³)")
ax4.set_ylabel("Trial score")
ax4.set_title("d) Density contrast estimation", fontsize=16)
ax4.set_yscale("log")
ax4.yaxis.set_major_formatter(mpl.ticker.StrMethodFormatter("{x:.0f}"))
plt.tight_layout()
fig.savefig(
"../paper/figures/Ross_Sea_inversion_results_subplots.png",
dpi=300,
bbox_inches="tight",
)